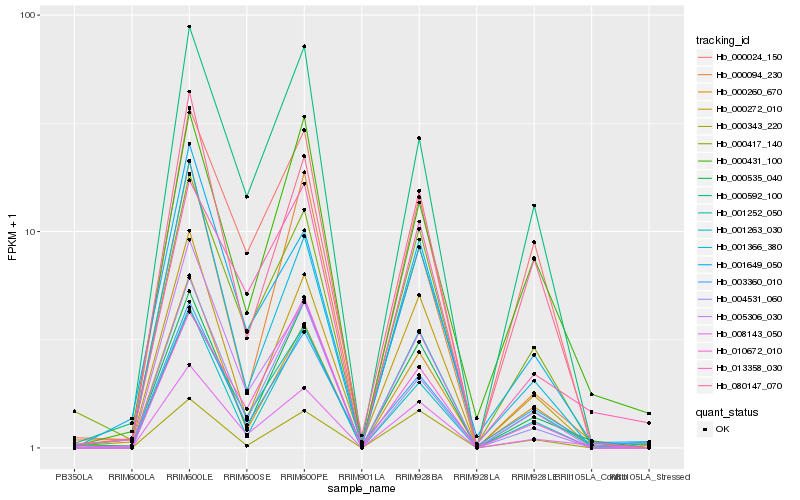
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008143_050 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 710A1-like [Jatropha curcas] |
| 2 |
Hb_001252_050 |
0.098852953 |
- |
- |
Wall-associated receptor kinase-like 20 [Morus notabilis] |
| 3 |
Hb_000535_040 |
0.1034003003 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 4 |
Hb_005306_030 |
0.1059083025 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 5 |
Hb_000417_140 |
0.1168704544 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 6 |
Hb_000272_010 |
0.1200455174 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 7 |
Hb_000260_670 |
0.1212140125 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 8 |
Hb_013358_030 |
0.1250787217 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 9 |
Hb_001366_380 |
0.1276460036 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_003360_010 |
0.1286345304 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 11 |
Hb_080147_070 |
0.1302102293 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 12 |
Hb_000592_100 |
0.1313660818 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 13 |
Hb_001263_030 |
0.1350200066 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 14 |
Hb_001649_050 |
0.1361081288 |
- |
- |
Shaggy-like kinase 13 isoform 1 [Theobroma cacao] |
| 15 |
Hb_000094_230 |
0.137733296 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |
| 16 |
Hb_000343_220 |
0.1389703207 |
- |
- |
hypothetical protein CISIN_1g037489mg [Citrus sinensis] |
| 17 |
Hb_004531_060 |
0.1460601537 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 18 |
Hb_000431_100 |
0.1480031051 |
- |
- |
ribonucleoside-diphosphate reductase small chain, putative [Ricinus communis] |
| 19 |
Hb_000024_150 |
0.149396584 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 20 |
Hb_010672_010 |
0.1500470213 |
- |
- |
hypothetical protein JCGZ_00895 [Jatropha curcas] |