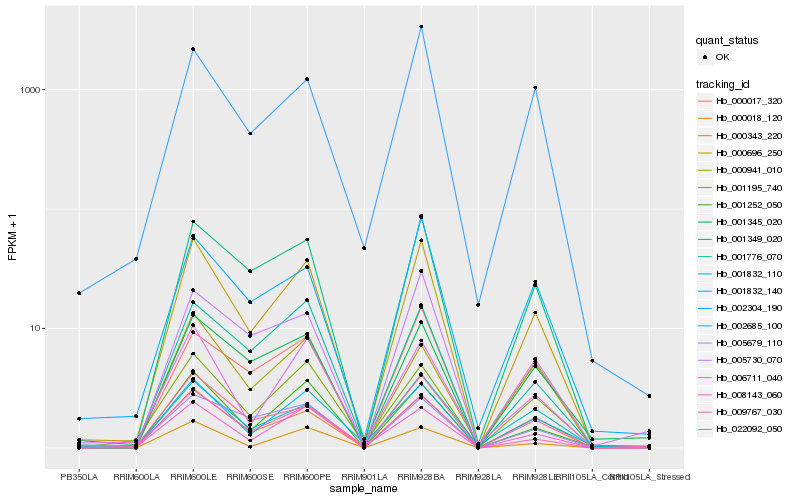
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000696_250 |
0.0 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 2 |
Hb_001832_140 |
0.0942654525 |
- |
- |
endomembrane protein 70 family protein [Medicago truncatula] |
| 3 |
Hb_000018_120 |
0.0990816276 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_001252_050 |
0.0990958842 |
- |
- |
Wall-associated receptor kinase-like 20 [Morus notabilis] |
| 5 |
Hb_001349_020 |
0.1004304542 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 6 |
Hb_009767_030 |
0.1060834495 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 7 |
Hb_022092_050 |
0.1109835188 |
- |
- |
PREDICTED: uncharacterized protein LOC105136214 [Populus euphratica] |
| 8 |
Hb_001195_740 |
0.1127274305 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006711_040 |
0.1144731658 |
- |
- |
PREDICTED: probable protein ABIL5 [Jatropha curcas] |
| 10 |
Hb_008143_060 |
0.1147230215 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 11 |
Hb_001832_110 |
0.1196921796 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000343_220 |
0.1259675007 |
- |
- |
hypothetical protein CISIN_1g037489mg [Citrus sinensis] |
| 13 |
Hb_001776_070 |
0.1278934138 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 14 |
Hb_000017_320 |
0.1292802169 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor GLABRA 3-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002304_190 |
0.129694254 |
- |
- |
PREDICTED: 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase [Jatropha curcas] |
| 16 |
Hb_001345_020 |
0.1306377749 |
- |
- |
hypothetical protein POPTR_0007s10400g [Populus trichocarpa] |
| 17 |
Hb_005679_110 |
0.1341719711 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 18 |
Hb_005730_070 |
0.1362766815 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 8 [Jatropha curcas] |
| 19 |
Hb_000941_010 |
0.1396502817 |
- |
- |
hypothetical protein RCOM_0504460 [Ricinus communis] |
| 20 |
Hb_002685_100 |
0.1396849302 |
- |
- |
low temperature inducible SLTI66 [Glycine max] |