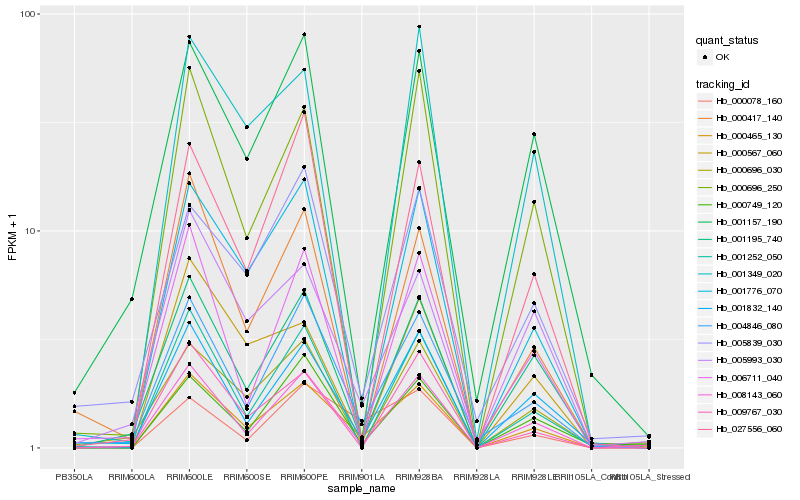
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_130 |
0.0 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0001s15550g [Populus trichocarpa] |
| 2 |
Hb_004846_080 |
0.1167033236 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 3 |
Hb_001832_140 |
0.1190512365 |
- |
- |
endomembrane protein 70 family protein [Medicago truncatula] |
| 4 |
Hb_008143_060 |
0.1204044648 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 5 |
Hb_001195_740 |
0.1368772227 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001776_070 |
0.13805066 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 7 |
Hb_001252_050 |
0.1416425713 |
- |
- |
Wall-associated receptor kinase-like 20 [Morus notabilis] |
| 8 |
Hb_000696_250 |
0.1451002383 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 9 |
Hb_027556_060 |
0.1570044998 |
- |
- |
hypothetical protein POPTR_0001s03520g [Populus trichocarpa] |
| 10 |
Hb_006711_040 |
0.1590271987 |
- |
- |
PREDICTED: probable protein ABIL5 [Jatropha curcas] |
| 11 |
Hb_005993_030 |
0.1598262048 |
- |
- |
putative senescence-associated protein SAG102 [Populus trichocarpa] |
| 12 |
Hb_000749_120 |
0.1622688208 |
- |
- |
PREDICTED: uncharacterized protein LOC105644255 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000417_140 |
0.1644173445 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 14 |
Hb_000696_030 |
0.1648824746 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |
| 15 |
Hb_009767_030 |
0.1677926685 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 16 |
Hb_001349_020 |
0.167833782 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 17 |
Hb_000078_160 |
0.1690697983 |
- |
- |
PREDICTED: protein LURP-one-related 12-like [Jatropha curcas] |
| 18 |
Hb_001157_190 |
0.1701012982 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 19 |
Hb_000567_060 |
0.1706040986 |
- |
- |
PREDICTED: U-box domain-containing protein 7 [Jatropha curcas] |
| 20 |
Hb_005839_030 |
0.1754883266 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |