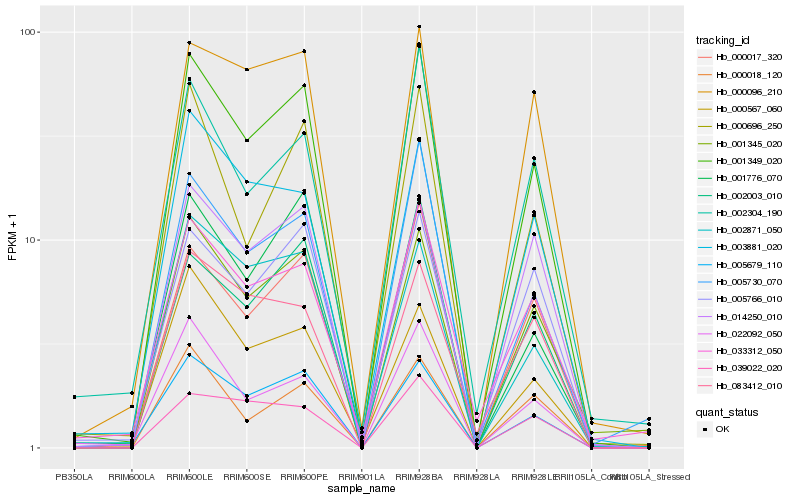
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001349_020 |
0.0 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 2 |
Hb_005679_110 |
0.0783082162 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 3 |
Hb_001345_020 |
0.0890518767 |
- |
- |
hypothetical protein POPTR_0007s10400g [Populus trichocarpa] |
| 4 |
Hb_000017_320 |
0.0909100548 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor GLABRA 3-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_005730_070 |
0.0909846723 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 8 [Jatropha curcas] |
| 6 |
Hb_000696_250 |
0.1004304542 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 7 |
Hb_002003_010 |
0.1041131122 |
- |
- |
hypothetical protein JCGZ_15062 [Jatropha curcas] |
| 8 |
Hb_005766_010 |
0.1109169751 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 9 |
Hb_033312_050 |
0.1126673986 |
- |
- |
PREDICTED: uncharacterized protein LOC105638598, partial [Jatropha curcas] |
| 10 |
Hb_001776_070 |
0.1126683632 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase isoform X2 [Jatropha curcas] |
| 11 |
Hb_022092_050 |
0.11468117 |
- |
- |
PREDICTED: uncharacterized protein LOC105136214 [Populus euphratica] |
| 12 |
Hb_002871_050 |
0.1154708199 |
transcription factor |
TF Family: ARF |
auxin response factor 1 family protein [Populus trichocarpa] |
| 13 |
Hb_000096_210 |
0.1171091942 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 14 |
Hb_014250_010 |
0.1174583402 |
- |
- |
PREDICTED: alpha-mannosidase-like isoform X2 [Populus euphratica] |
| 15 |
Hb_000018_120 |
0.1188567739 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_002304_190 |
0.1189982268 |
- |
- |
PREDICTED: 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase [Jatropha curcas] |
| 17 |
Hb_000567_060 |
0.1211702589 |
- |
- |
PREDICTED: U-box domain-containing protein 7 [Jatropha curcas] |
| 18 |
Hb_003881_020 |
0.1213205441 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 19 |
Hb_039022_020 |
0.1233115847 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 20 |
Hb_083412_010 |
0.1303298265 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |