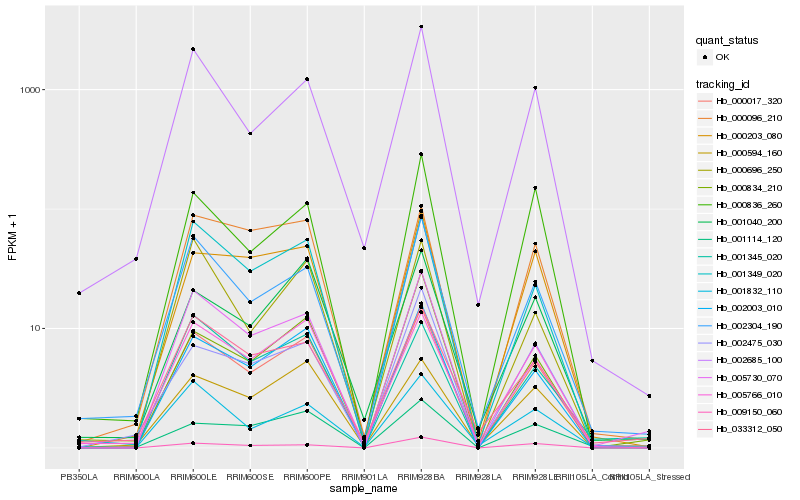
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000017_320 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor GLABRA 3-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002003_010 |
0.0658239332 |
- |
- |
hypothetical protein JCGZ_15062 [Jatropha curcas] |
| 3 |
Hb_001349_020 |
0.0909100548 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 4 |
Hb_002304_190 |
0.0957218776 |
- |
- |
PREDICTED: 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase [Jatropha curcas] |
| 5 |
Hb_005766_010 |
0.0992463536 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 6 |
Hb_005730_070 |
0.1078970152 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 8 [Jatropha curcas] |
| 7 |
Hb_002685_100 |
0.1111478018 |
- |
- |
low temperature inducible SLTI66 [Glycine max] |
| 8 |
Hb_000594_160 |
0.1130735631 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 9 |
Hb_000203_080 |
0.1155685085 |
- |
- |
Xylem bark cysteine peptidase 3 isoform 1 [Theobroma cacao] |
| 10 |
Hb_033312_050 |
0.1195446186 |
- |
- |
PREDICTED: uncharacterized protein LOC105638598, partial [Jatropha curcas] |
| 11 |
Hb_001114_120 |
0.1218217398 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_001832_110 |
0.1227607979 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000834_210 |
0.1230126806 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_001040_200 |
0.125424004 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 15 |
Hb_000096_210 |
0.1266543731 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 16 |
Hb_009150_060 |
0.1281906673 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |
| 17 |
Hb_000836_260 |
0.1284993299 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 18 |
Hb_002475_030 |
0.1288264278 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000696_250 |
0.1292802169 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 20 |
Hb_001345_020 |
0.1315275387 |
- |
- |
hypothetical protein POPTR_0007s10400g [Populus trichocarpa] |