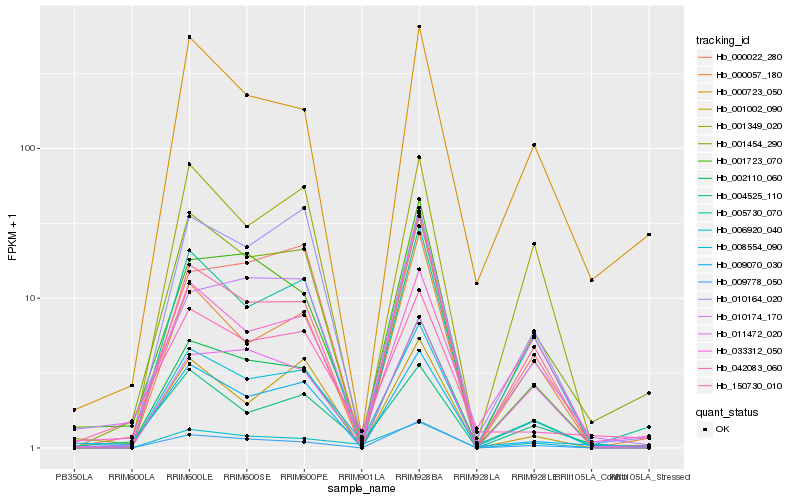
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008554_090 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_002110_060 |
0.0885334411 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 3 |
Hb_005730_070 |
0.098538151 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 8 [Jatropha curcas] |
| 4 |
Hb_009070_030 |
0.1201800688 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 5 |
Hb_004525_110 |
0.1258139744 |
- |
- |
hypothetical protein POPTR_0007s12850g [Populus trichocarpa] |
| 6 |
Hb_150730_010 |
0.1271181037 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 7 |
Hb_042083_060 |
0.1335923293 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 8 |
Hb_001454_290 |
0.1345606538 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 9 |
Hb_033312_050 |
0.1361579599 |
- |
- |
PREDICTED: uncharacterized protein LOC105638598, partial [Jatropha curcas] |
| 10 |
Hb_001349_020 |
0.1366193818 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 11 |
Hb_000057_180 |
0.1376663086 |
- |
- |
hypothetical protein POPTR_0001s18200g [Populus trichocarpa] |
| 12 |
Hb_006920_040 |
0.1399232592 |
- |
- |
PREDICTED: uncharacterized protein LOC103341644 [Prunus mume] |
| 13 |
Hb_009778_050 |
0.1411225101 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 14 |
Hb_000723_050 |
0.1421570658 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 15 |
Hb_000022_280 |
0.1430805161 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 16 |
Hb_001002_090 |
0.1435932057 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 17 |
Hb_010174_170 |
0.1440285689 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001723_070 |
0.1490863613 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 19 |
Hb_010164_020 |
0.1497412024 |
- |
- |
PREDICTED: abscisate beta-glucosyltransferase-like [Jatropha curcas] |
| 20 |
Hb_011472_020 |
0.1530991194 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |