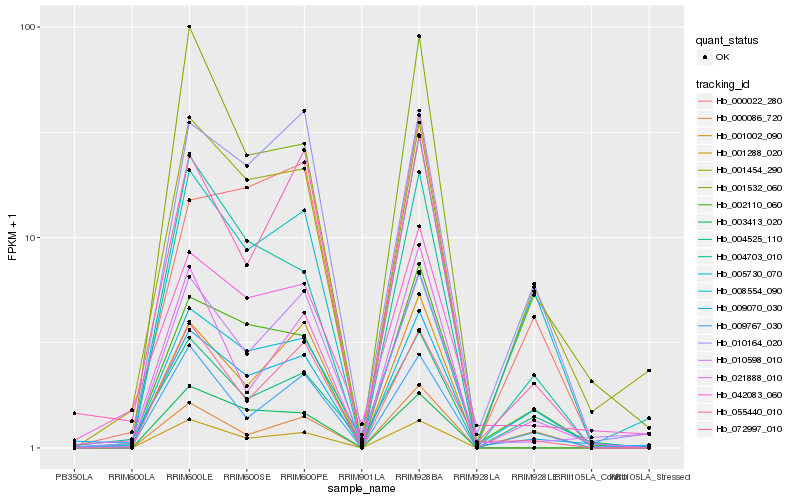
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009070_030 |
0.0 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 2 |
Hb_042083_060 |
0.1103485489 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 3 |
Hb_008554_090 |
0.1201800688 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_002110_060 |
0.1202379405 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 5 |
Hb_010598_010 |
0.1297396018 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 6 |
Hb_001002_090 |
0.1305911424 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 7 |
Hb_055440_010 |
0.1311307814 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_001454_290 |
0.1432606395 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 9 |
Hb_001288_020 |
0.1460883719 |
- |
- |
- |
| 10 |
Hb_021888_010 |
0.1521734276 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_072997_010 |
0.1529133371 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 12 |
Hb_004525_110 |
0.1538836251 |
- |
- |
hypothetical protein POPTR_0007s12850g [Populus trichocarpa] |
| 13 |
Hb_005730_070 |
0.1593990333 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 8 [Jatropha curcas] |
| 14 |
Hb_010164_020 |
0.160351808 |
- |
- |
PREDICTED: abscisate beta-glucosyltransferase-like [Jatropha curcas] |
| 15 |
Hb_001532_060 |
0.1611298471 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_009767_030 |
0.1626641438 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 17 |
Hb_003413_020 |
0.1644357474 |
- |
- |
- |
| 18 |
Hb_000086_720 |
0.1647608032 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 19 |
Hb_004703_010 |
0.1734430649 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 20 |
Hb_000022_280 |
0.1754569879 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |