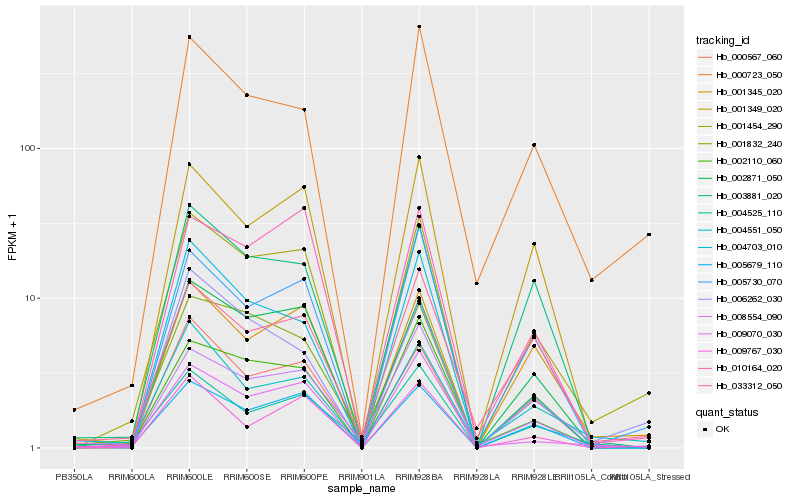
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001454_290 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 2 |
Hb_002871_050 |
0.1092899548 |
transcription factor |
TF Family: ARF |
auxin response factor 1 family protein [Populus trichocarpa] |
| 3 |
Hb_005679_110 |
0.1247161098 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 4 |
Hb_001345_020 |
0.1248724628 |
- |
- |
hypothetical protein POPTR_0007s10400g [Populus trichocarpa] |
| 5 |
Hb_005730_070 |
0.126302362 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 8 [Jatropha curcas] |
| 6 |
Hb_002110_060 |
0.128264052 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 7 |
Hb_000723_050 |
0.1289876889 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 8 |
Hb_000567_060 |
0.1334619602 |
- |
- |
PREDICTED: U-box domain-containing protein 7 [Jatropha curcas] |
| 9 |
Hb_008554_090 |
0.1345606538 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_001832_240 |
0.1357900669 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 11 |
Hb_001349_020 |
0.1366084647 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 12 |
Hb_010164_020 |
0.1393828068 |
- |
- |
PREDICTED: abscisate beta-glucosyltransferase-like [Jatropha curcas] |
| 13 |
Hb_009767_030 |
0.1394616678 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 14 |
Hb_004551_050 |
0.1405302216 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_009070_030 |
0.1432606395 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 16 |
Hb_006262_030 |
0.1441723179 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_003881_020 |
0.1475391902 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 18 |
Hb_004525_110 |
0.1483977147 |
- |
- |
hypothetical protein POPTR_0007s12850g [Populus trichocarpa] |
| 19 |
Hb_033312_050 |
0.1491399244 |
- |
- |
PREDICTED: uncharacterized protein LOC105638598, partial [Jatropha curcas] |
| 20 |
Hb_004703_010 |
0.1540475053 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |