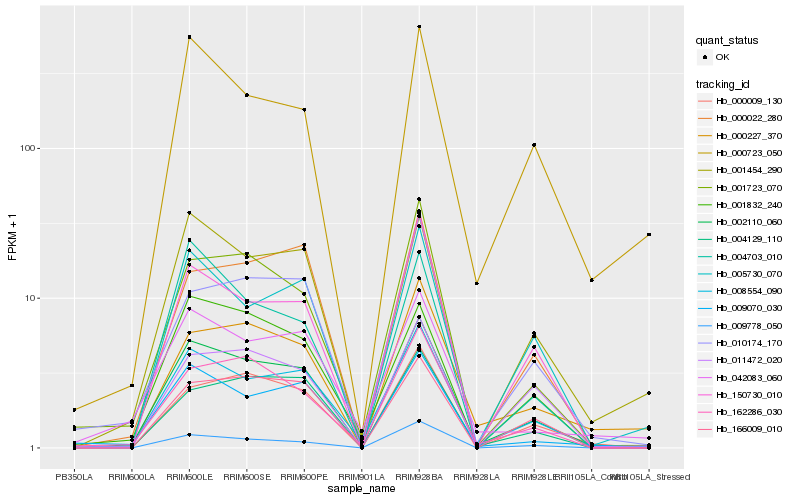
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002110_060 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 2 |
Hb_008554_090 |
0.0885334411 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_009070_030 |
0.1202379405 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 4 |
Hb_162286_030 |
0.1221554902 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 5 |
Hb_000009_130 |
0.1233611667 |
- |
- |
hypothetical protein POPTR_0007s12410g [Populus trichocarpa] |
| 6 |
Hb_005730_070 |
0.1249132713 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 8 [Jatropha curcas] |
| 7 |
Hb_001454_290 |
0.128264052 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 8 |
Hb_150730_010 |
0.1288299861 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 9 |
Hb_004129_110 |
0.1292069391 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 10 |
Hb_166009_010 |
0.1292208778 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 11 |
Hb_001723_070 |
0.130220575 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 12 |
Hb_009778_050 |
0.1336224323 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 13 |
Hb_000022_280 |
0.1369838503 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 14 |
Hb_001832_240 |
0.1375026107 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 15 |
Hb_011472_020 |
0.1417241861 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |
| 16 |
Hb_000723_050 |
0.1424584267 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 17 |
Hb_010174_170 |
0.1435295042 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000227_370 |
0.145107453 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 19 |
Hb_004703_010 |
0.1456799061 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 20 |
Hb_042083_060 |
0.1521086389 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |