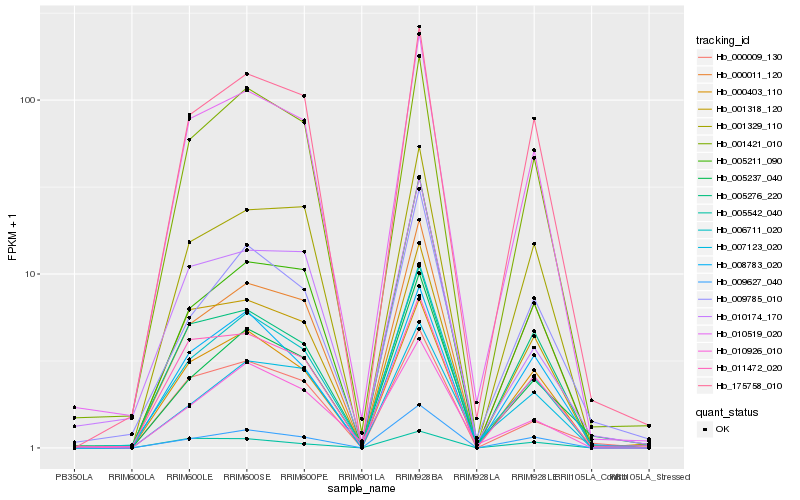
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010519_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_19028 [Jatropha curcas] |
| 2 |
Hb_001318_120 |
0.0597183409 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 3 |
Hb_175758_010 |
0.0649726801 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_001421_010 |
0.0665113027 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_011472_020 |
0.0728464434 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |
| 6 |
Hb_006711_020 |
0.0773524046 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |
| 7 |
Hb_000403_110 |
0.0807213665 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 8 |
Hb_005276_220 |
0.0856224415 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 9 |
Hb_001329_110 |
0.0857812792 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 10 |
Hb_000011_120 |
0.0937352352 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 11 |
Hb_008783_020 |
0.1053645296 |
- |
- |
hypothetical protein JCGZ_21284 [Jatropha curcas] |
| 12 |
Hb_009627_040 |
0.1101677986 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 13 |
Hb_000009_130 |
0.1127269047 |
- |
- |
hypothetical protein POPTR_0007s12410g [Populus trichocarpa] |
| 14 |
Hb_009785_010 |
0.1142731724 |
- |
- |
PREDICTED: uncharacterized protein LOC105648749 [Jatropha curcas] |
| 15 |
Hb_005211_090 |
0.1152079599 |
- |
- |
PREDICTED: aluminum-activated malate transporter 4-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_005237_040 |
0.1158897391 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 17 |
Hb_007123_020 |
0.1183912217 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HOX3 [Jatropha curcas] |
| 18 |
Hb_010174_170 |
0.1193677074 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005542_040 |
0.1194115852 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 20 |
Hb_010926_010 |
0.1194117135 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |