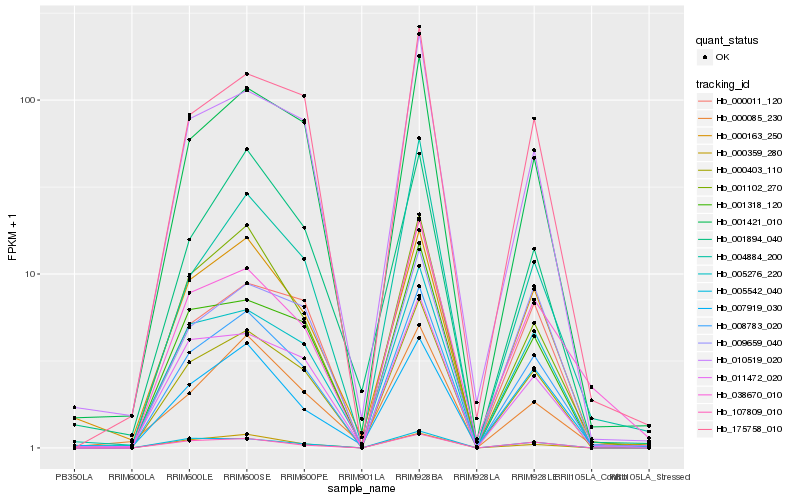
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008783_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_21284 [Jatropha curcas] |
| 2 |
Hb_000403_110 |
0.0705658988 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 3 |
Hb_107809_010 |
0.0849118877 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 4 |
Hb_005276_220 |
0.0884718983 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 5 |
Hb_001102_270 |
0.0887525935 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 6 |
Hb_001421_010 |
0.0910394553 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_175758_010 |
0.0922212083 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_001318_120 |
0.1016270923 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 9 |
Hb_010519_020 |
0.1053645296 |
- |
- |
hypothetical protein JCGZ_19028 [Jatropha curcas] |
| 10 |
Hb_005542_040 |
0.1054063797 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 11 |
Hb_000163_250 |
0.1057217881 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_000359_280 |
0.1057640617 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |
| 13 |
Hb_011472_020 |
0.1098634743 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |
| 14 |
Hb_000011_120 |
0.1159106525 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 15 |
Hb_038670_010 |
0.1214444029 |
- |
- |
Major allergen Pru ar, putative [Ricinus communis] |
| 16 |
Hb_009659_040 |
0.1222760098 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER [Populus euphratica] |
| 17 |
Hb_007919_030 |
0.1232358961 |
- |
- |
- |
| 18 |
Hb_004884_200 |
0.124690442 |
- |
- |
PREDICTED: ferritin-3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001894_040 |
0.127603914 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000085_230 |
0.1300098681 |
- |
- |
PREDICTED: probable long-chain-alcohol O-fatty-acyltransferase 5 [Jatropha curcas] |