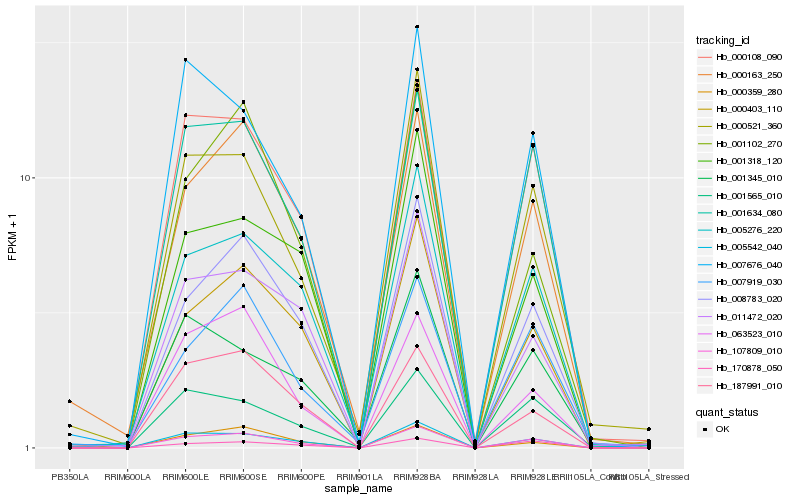
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_107809_010 |
0.0 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 2 |
Hb_005542_040 |
0.0606392272 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 3 |
Hb_001634_080 |
0.0784713362 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_008783_020 |
0.0849118877 |
- |
- |
hypothetical protein JCGZ_21284 [Jatropha curcas] |
| 5 |
Hb_000108_090 |
0.0864466163 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 6 |
Hb_005276_220 |
0.096460991 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 7 |
Hb_000521_360 |
0.100537626 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 8 |
Hb_000403_110 |
0.1057229174 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 9 |
Hb_007676_040 |
0.1095435726 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 10 |
Hb_000359_280 |
0.1104499222 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |
| 11 |
Hb_170878_050 |
0.1131083508 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 12 |
Hb_000163_250 |
0.1149323631 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_001102_270 |
0.1169898816 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 14 |
Hb_063523_010 |
0.1180532623 |
- |
- |
- |
| 15 |
Hb_187991_010 |
0.1195196481 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 16 |
Hb_001318_120 |
0.1208175174 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 17 |
Hb_007919_030 |
0.1252825662 |
- |
- |
- |
| 18 |
Hb_011472_020 |
0.1258250893 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |
| 19 |
Hb_001565_010 |
0.1265519683 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 20 |
Hb_001345_010 |
0.1341905879 |
- |
- |
hypothetical protein MIMGU_mgv1a010889mg [Erythranthe guttata] |