| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
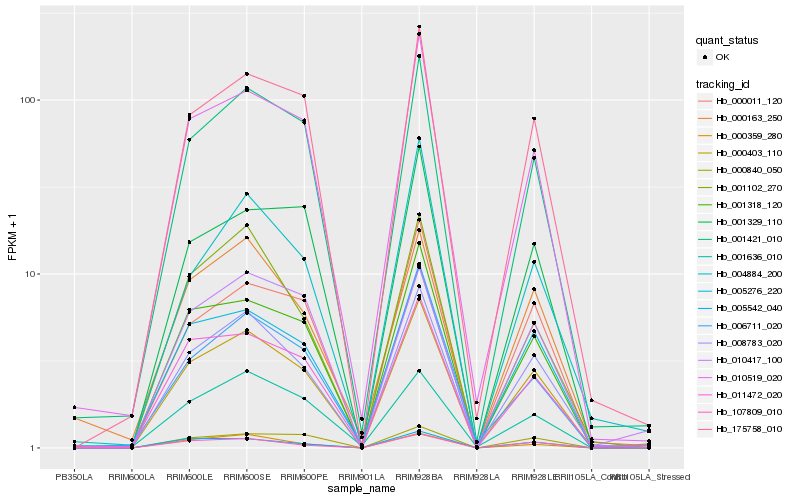
Hb_000403_110 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 2 |
Hb_008783_020 |
0.0705658988 |
- |
- |
hypothetical protein JCGZ_21284 [Jatropha curcas] |
| 3 |
Hb_001421_010 |
0.0717614354 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005276_220 |
0.0726618284 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 5 |
Hb_010519_020 |
0.0807213665 |
- |
- |
hypothetical protein JCGZ_19028 [Jatropha curcas] |
| 6 |
Hb_175758_010 |
0.0808594559 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_001318_120 |
0.0836090569 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 8 |
Hb_011472_020 |
0.0845092647 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |
| 9 |
Hb_000011_120 |
0.100635087 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 10 |
Hb_107809_010 |
0.1057229174 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 11 |
Hb_005542_040 |
0.1063564144 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 12 |
Hb_000163_250 |
0.1119238011 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_006711_020 |
0.1119298278 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |
| 14 |
Hb_001329_110 |
0.1149221327 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 15 |
Hb_001102_270 |
0.1171881885 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 16 |
Hb_000840_050 |
0.1179820436 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 17 |
Hb_000359_280 |
0.1214237332 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |
| 18 |
Hb_001636_010 |
0.1215740033 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 19 |
Hb_010417_100 |
0.1227477098 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 20 |
Hb_004884_200 |
0.1246773793 |
- |
- |
PREDICTED: ferritin-3, chloroplastic [Jatropha curcas] |