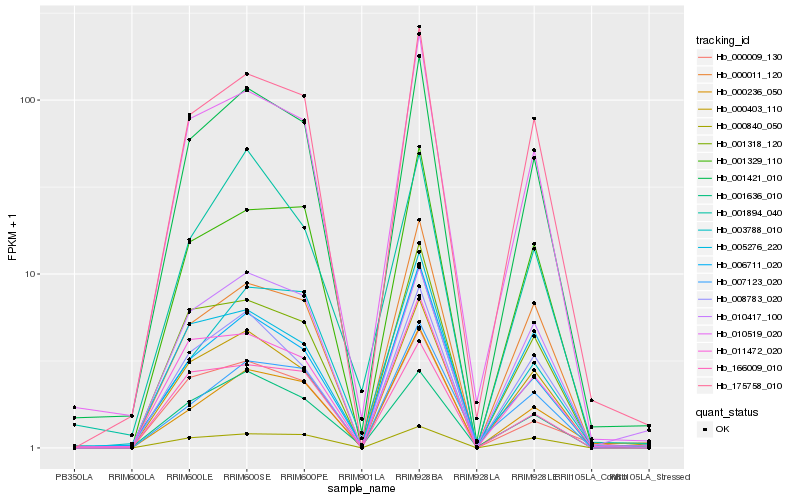
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001421_010 |
0.0 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_175758_010 |
0.0528134672 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_010519_020 |
0.0665113027 |
- |
- |
hypothetical protein JCGZ_19028 [Jatropha curcas] |
| 4 |
Hb_000403_110 |
0.0717614354 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 5 |
Hb_011472_020 |
0.0832871352 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |
| 6 |
Hb_001329_110 |
0.0910232822 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 7 |
Hb_008783_020 |
0.0910394553 |
- |
- |
hypothetical protein JCGZ_21284 [Jatropha curcas] |
| 8 |
Hb_001318_120 |
0.0943089226 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 9 |
Hb_005276_220 |
0.0956507073 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 10 |
Hb_001636_010 |
0.097726577 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 11 |
Hb_000840_050 |
0.0985576445 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 12 |
Hb_006711_020 |
0.1020763272 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |
| 13 |
Hb_010417_100 |
0.103214857 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 14 |
Hb_000011_120 |
0.1071167571 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 15 |
Hb_166009_010 |
0.1113819903 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 16 |
Hb_001894_040 |
0.1143385799 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007123_020 |
0.1151985019 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HOX3 [Jatropha curcas] |
| 18 |
Hb_000236_050 |
0.1156435024 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 19 |
Hb_000009_130 |
0.1156795855 |
- |
- |
hypothetical protein POPTR_0007s12410g [Populus trichocarpa] |
| 20 |
Hb_003788_010 |
0.1160969634 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |