| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
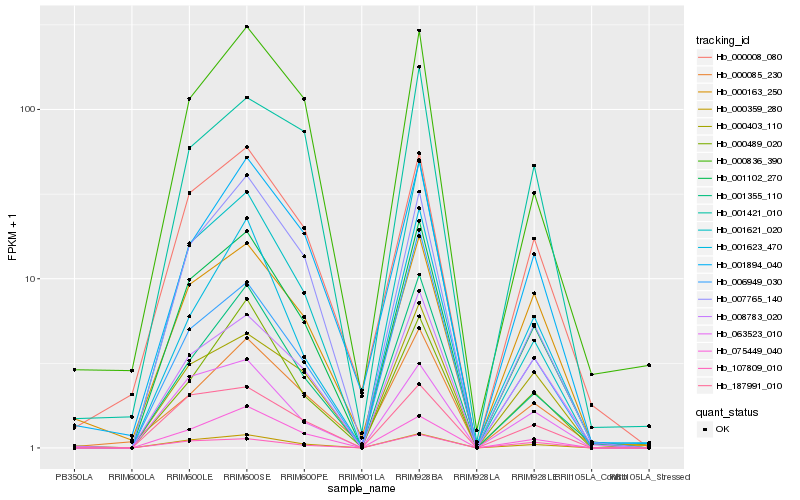
Hb_001102_270 |
0.0 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 2 |
Hb_000359_280 |
0.0447551327 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |
| 3 |
Hb_008783_020 |
0.0887525935 |
- |
- |
hypothetical protein JCGZ_21284 [Jatropha curcas] |
| 4 |
Hb_001355_110 |
0.09471336 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_187991_010 |
0.0995981342 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 6 |
Hb_063523_010 |
0.0998092969 |
- |
- |
- |
| 7 |
Hb_007765_140 |
0.1075448564 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |
| 8 |
Hb_001621_020 |
0.1090337718 |
- |
- |
- |
| 9 |
Hb_000008_080 |
0.1106349467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000489_020 |
0.1167124542 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 11 |
Hb_107809_010 |
0.1169898816 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 12 |
Hb_000403_110 |
0.1171881885 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 13 |
Hb_075449_040 |
0.1200389958 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-1 [Jatropha curcas] |
| 14 |
Hb_001894_040 |
0.1224370432 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000085_230 |
0.1247996726 |
- |
- |
PREDICTED: probable long-chain-alcohol O-fatty-acyltransferase 5 [Jatropha curcas] |
| 16 |
Hb_000836_390 |
0.1258257262 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 17 |
Hb_000163_250 |
0.1275533968 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_001623_470 |
0.1276319272 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 19 |
Hb_001421_010 |
0.1280391209 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006949_030 |
0.1309450482 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |