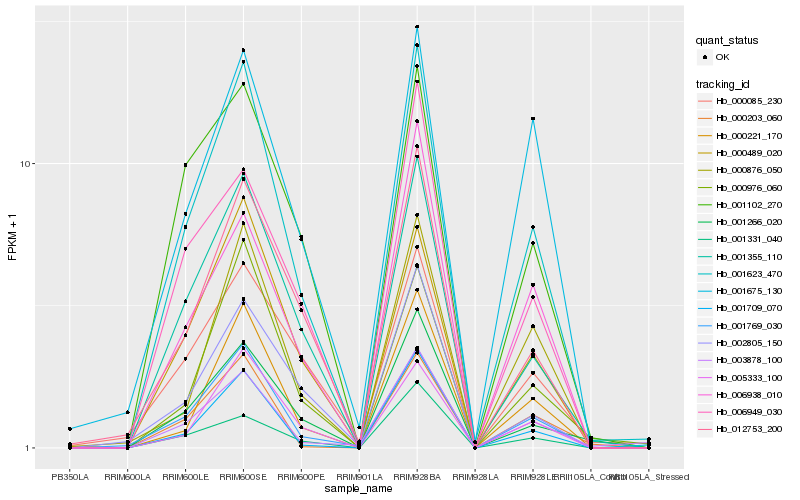
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001623_470 |
0.0 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 2 |
Hb_001355_110 |
0.0905009646 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001769_030 |
0.0925195032 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 4 |
Hb_000489_020 |
0.1041972288 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 5 |
Hb_005333_100 |
0.1173501794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001266_020 |
0.1192389876 |
- |
- |
PREDICTED: uncharacterized protein LOC105650395 [Jatropha curcas] |
| 7 |
Hb_000976_060 |
0.1214413725 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000221_170 |
0.124976034 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 2 [Jatropha curcas] |
| 9 |
Hb_012753_200 |
0.1264368836 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |
| 10 |
Hb_001709_070 |
0.1274636232 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 11 |
Hb_001102_270 |
0.1276319272 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 12 |
Hb_003878_100 |
0.1278722436 |
desease resistance |
Gene Name: NB-ARC |
LRR and NB-ARC domains-containing disease resistance protein, putative [Theobroma cacao] |
| 13 |
Hb_006949_030 |
0.1363643736 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000876_050 |
0.1379265084 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 15 |
Hb_001331_040 |
0.1384529359 |
- |
- |
PREDICTED: cationic peroxidase 1-like [Jatropha curcas] |
| 16 |
Hb_000203_060 |
0.139546311 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0004s05660g [Populus trichocarpa] |
| 17 |
Hb_006938_010 |
0.1398449281 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 18 |
Hb_002805_150 |
0.1403839996 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 19 |
Hb_001675_130 |
0.1409131051 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 20 |
Hb_000085_230 |
0.1427035161 |
- |
- |
PREDICTED: probable long-chain-alcohol O-fatty-acyltransferase 5 [Jatropha curcas] |