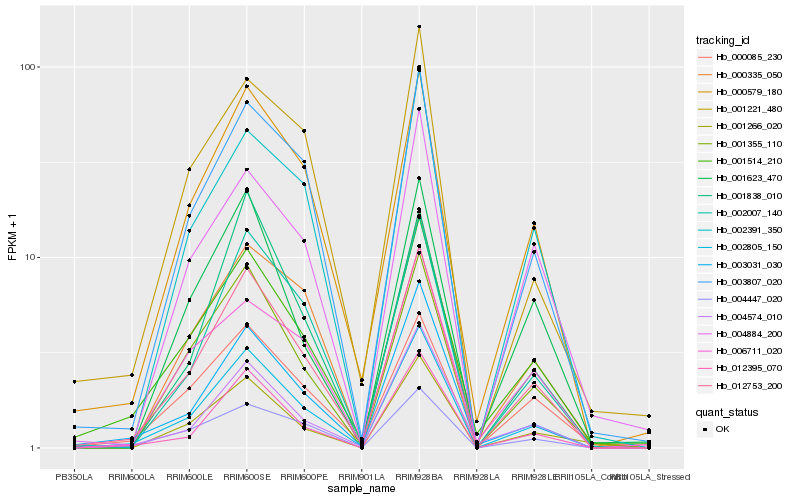
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012753_200 |
0.0 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |
| 2 |
Hb_002805_150 |
0.0378961503 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 3 |
Hb_000579_180 |
0.0761263804 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 4 |
Hb_003807_020 |
0.0868194961 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 5 |
Hb_002007_140 |
0.0894766421 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like [Populus euphratica] |
| 6 |
Hb_012395_070 |
0.0919189203 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL6 [Jatropha curcas] |
| 7 |
Hb_001514_210 |
0.0923872605 |
- |
- |
hypothetical protein VITISV_012133 [Vitis vinifera] |
| 8 |
Hb_001355_110 |
0.097743406 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_004447_020 |
0.1045838883 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 10 |
Hb_003031_030 |
0.1094383618 |
- |
- |
hypothetical protein JCGZ_21288 [Jatropha curcas] |
| 11 |
Hb_000085_230 |
0.1099485247 |
- |
- |
PREDICTED: probable long-chain-alcohol O-fatty-acyltransferase 5 [Jatropha curcas] |
| 12 |
Hb_000335_050 |
0.1123848087 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002391_350 |
0.1169208016 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |
| 14 |
Hb_004574_010 |
0.1184343038 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 15 |
Hb_001266_020 |
0.1201164837 |
- |
- |
PREDICTED: uncharacterized protein LOC105650395 [Jatropha curcas] |
| 16 |
Hb_001623_470 |
0.1264368836 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 17 |
Hb_001221_480 |
0.1275948486 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_004884_200 |
0.128636204 |
- |
- |
PREDICTED: ferritin-3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001838_010 |
0.1287571965 |
- |
- |
hypothetical protein POPTR_0005s18380g [Populus trichocarpa] |
| 20 |
Hb_006711_020 |
0.1317881261 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |