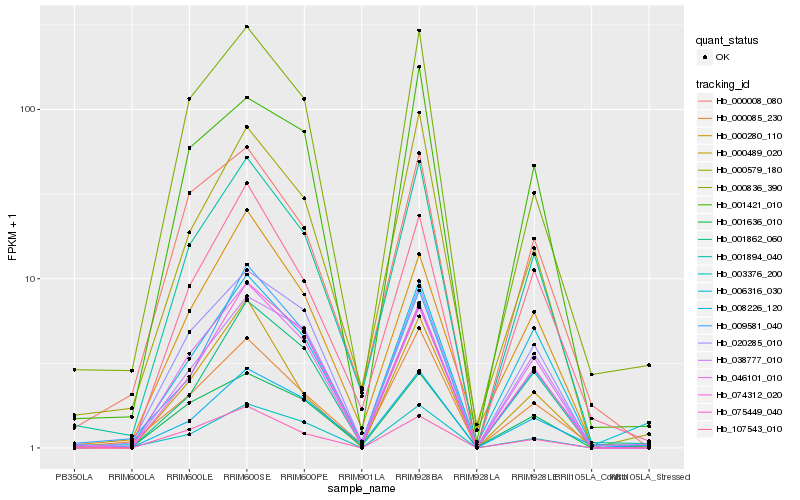
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001894_040 |
0.0 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000085_230 |
0.0835463849 |
- |
- |
PREDICTED: probable long-chain-alcohol O-fatty-acyltransferase 5 [Jatropha curcas] |
| 3 |
Hb_008226_120 |
0.0873191183 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_000579_180 |
0.0876438572 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 5 |
Hb_046101_010 |
0.08999334 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 6 |
Hb_001862_060 |
0.0926125666 |
- |
- |
PREDICTED: uncharacterized protein LOC105639802 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001636_010 |
0.1018370106 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 8 |
Hb_000008_080 |
0.1023973188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_107543_010 |
0.1039492128 |
- |
- |
Cysteine-rich RLK 10, putative [Theobroma cacao] |
| 10 |
Hb_074312_020 |
0.104687999 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_038777_010 |
0.1049827418 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |
| 12 |
Hb_009581_040 |
0.1051266721 |
- |
- |
hypothetical protein B456_011G052100 [Gossypium raimondii] |
| 13 |
Hb_020285_010 |
0.1058609715 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3, partial [Pyrus x bretschneideri] |
| 14 |
Hb_075449_040 |
0.1136151395 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-1 [Jatropha curcas] |
| 15 |
Hb_000836_390 |
0.1143043236 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 16 |
Hb_001421_010 |
0.1143385799 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000280_110 |
0.1145265583 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 18 |
Hb_003376_200 |
0.1167680901 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_006316_030 |
0.1167935561 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 20 |
Hb_000489_020 |
0.1174060789 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |