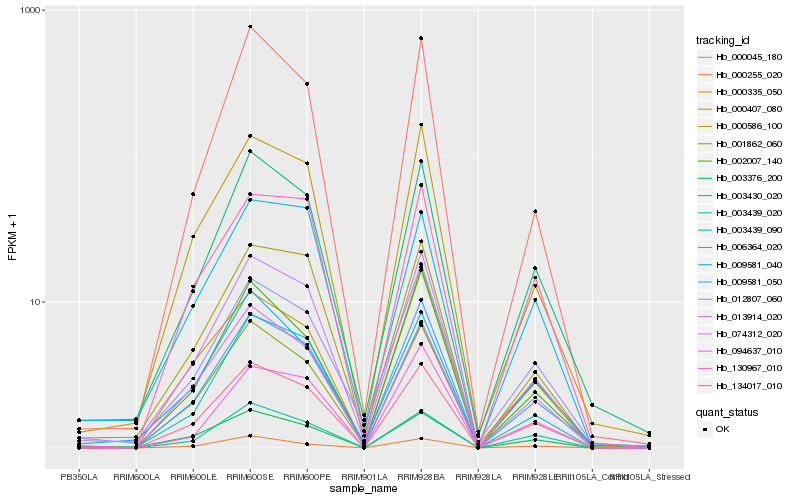
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_134017_010 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_003439_020 |
0.0733178704 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1-like [Solanum lycopersicum] |
| 3 |
Hb_003376_200 |
0.0757846848 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_003439_090 |
0.0783193151 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_074312_020 |
0.0848503749 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_130967_010 |
0.0854513289 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_003430_020 |
0.0857854915 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_009581_050 |
0.0872168523 |
- |
- |
hypothetical protein CISIN_1g0025812mg, partial [Citrus sinensis] |
| 9 |
Hb_001862_060 |
0.0876906799 |
- |
- |
PREDICTED: uncharacterized protein LOC105639802 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000407_080 |
0.0882577796 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase [Hevea brasiliensis] |
| 11 |
Hb_000255_020 |
0.0946248419 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 12 |
Hb_013914_020 |
0.1005795713 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 13 |
Hb_000586_100 |
0.1063937673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000335_050 |
0.1090709051 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002007_140 |
0.1123006875 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like [Populus euphratica] |
| 16 |
Hb_000045_180 |
0.1126663174 |
- |
- |
aquaporin PIP1 [Hevea brasiliensis] |
| 17 |
Hb_012807_060 |
0.1165822709 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_009581_040 |
0.1192226065 |
- |
- |
hypothetical protein B456_011G052100 [Gossypium raimondii] |
| 19 |
Hb_006364_020 |
0.1192666715 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |
| 20 |
Hb_094637_010 |
0.1195779994 |
- |
- |
PREDICTED: uncharacterized protein LOC105642042 [Jatropha curcas] |