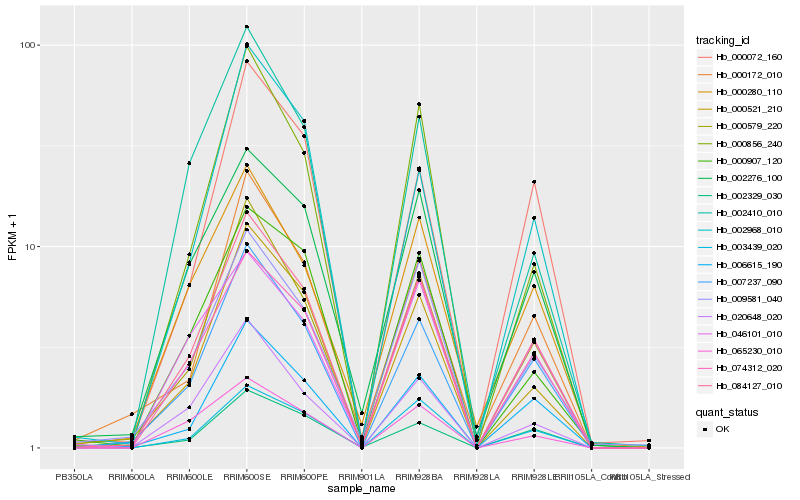
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_084127_010 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_007237_090 |
0.0495991847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000579_220 |
0.0650341719 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_006615_190 |
0.0729887988 |
- |
- |
UDP-glycosyltransferase 1 [Linum usitatissimum] |
| 5 |
Hb_000521_210 |
0.0877461954 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 6 |
Hb_074312_020 |
0.0917816298 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_020648_020 |
0.0933013501 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 8 |
Hb_003439_020 |
0.0969658804 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1-like [Solanum lycopersicum] |
| 9 |
Hb_002276_100 |
0.0981962946 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89A2-like [Jatropha curcas] |
| 10 |
Hb_002329_030 |
0.1029272243 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_009581_040 |
0.1030556061 |
- |
- |
hypothetical protein B456_011G052100 [Gossypium raimondii] |
| 12 |
Hb_000280_110 |
0.1074644988 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 13 |
Hb_000856_240 |
0.1107100375 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_065230_010 |
0.1115073787 |
- |
- |
PREDICTED: uncharacterized protein LOC100814166 isoform X1 [Glycine max] |
| 15 |
Hb_000072_160 |
0.1117383992 |
- |
- |
proline oxidase [Manihot esculenta] |
| 16 |
Hb_000172_010 |
0.1126514613 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 17 |
Hb_000907_120 |
0.1148751188 |
transcription factor |
TF Family: G2-like |
PREDICTED: transcription repressor KAN1-like [Populus euphratica] |
| 18 |
Hb_002410_010 |
0.116862614 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 19 |
Hb_002968_010 |
0.1181503835 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 20 |
Hb_046101_010 |
0.1212538706 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |