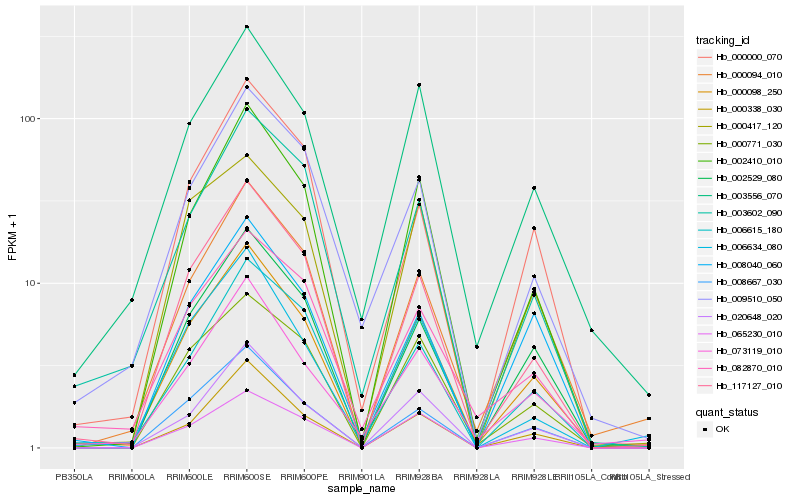
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000098_250 |
0.0 |
- |
- |
UDP-glucose flavonoid 3-O-glucosyltransferase [Medicago truncatula] |
| 2 |
Hb_002410_010 |
0.068642066 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 3 |
Hb_117127_010 |
0.0694269587 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH106 [Jatropha curcas] |
| 4 |
Hb_008667_030 |
0.0790582075 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 5 |
Hb_002529_080 |
0.0814656842 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase [Jatropha curcas] |
| 6 |
Hb_020648_020 |
0.0849591068 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 7 |
Hb_000771_030 |
0.0885704405 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 8 |
Hb_065230_010 |
0.0904669083 |
- |
- |
PREDICTED: uncharacterized protein LOC100814166 isoform X1 [Glycine max] |
| 9 |
Hb_000338_030 |
0.0931145447 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_21254 [Jatropha curcas] |
| 10 |
Hb_003556_070 |
0.0991762528 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 11 |
Hb_006615_180 |
0.1082265733 |
- |
- |
hypothetical protein B456_003G103400 [Gossypium raimondii] |
| 12 |
Hb_073119_010 |
0.108398032 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 13 |
Hb_000417_120 |
0.1095552588 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 14 |
Hb_000094_010 |
0.1122640294 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 15 |
Hb_000000_070 |
0.1173012999 |
- |
- |
allene oxide synthase [Hevea brasiliensis] |
| 16 |
Hb_003602_090 |
0.1179062963 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 17 |
Hb_006634_080 |
0.1194843533 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 18 |
Hb_008040_060 |
0.1196518335 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 19 |
Hb_082870_010 |
0.1201033385 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 20 |
Hb_009510_050 |
0.1203873854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |