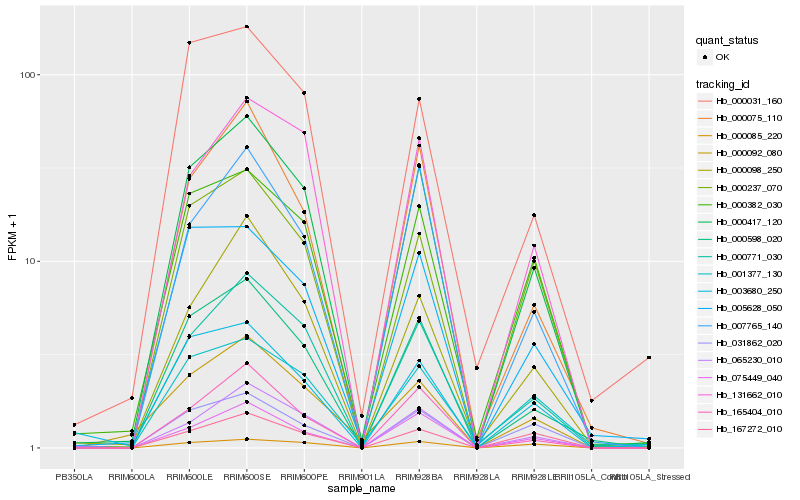
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_120 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 2 |
Hb_000771_030 |
0.0734108854 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000598_020 |
0.0772741326 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 4 |
Hb_065230_010 |
0.0941217446 |
- |
- |
PREDICTED: uncharacterized protein LOC100814166 isoform X1 [Glycine max] |
| 5 |
Hb_000382_030 |
0.0995264533 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 6 |
Hb_000237_070 |
0.1010053684 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 7 |
Hb_000075_110 |
0.1062202159 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 8 |
Hb_000098_250 |
0.1095552588 |
- |
- |
UDP-glucose flavonoid 3-O-glucosyltransferase [Medicago truncatula] |
| 9 |
Hb_131662_010 |
0.1105421396 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 10 |
Hb_005628_050 |
0.1142310755 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 11 |
Hb_007765_140 |
0.1173931459 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |
| 12 |
Hb_031862_020 |
0.1179952438 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 13 |
Hb_001377_130 |
0.1194550773 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 14 |
Hb_075449_040 |
0.1200973079 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-1 [Jatropha curcas] |
| 15 |
Hb_165404_010 |
0.1226315671 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 16 |
Hb_000031_160 |
0.1241404377 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 17 |
Hb_003680_250 |
0.1316535022 |
- |
- |
PREDICTED: probable 3-deoxy-D-manno-octulosonic acid transferase, mitochondrial isoform X2 [Jatropha curcas] |
| 18 |
Hb_000092_080 |
0.132308794 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_038742 [Vitis vinifera] |
| 19 |
Hb_000085_220 |
0.1327238685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_167272_010 |
0.1327362845 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |