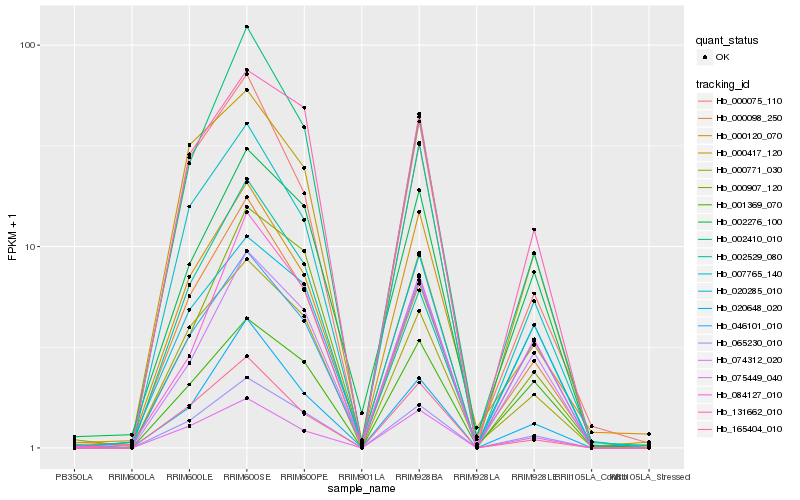
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065230_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100814166 isoform X1 [Glycine max] |
| 2 |
Hb_000771_030 |
0.0636791559 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 3 |
Hb_131662_010 |
0.0712042454 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 4 |
Hb_002410_010 |
0.0853397971 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 5 |
Hb_000098_250 |
0.0904669083 |
- |
- |
UDP-glucose flavonoid 3-O-glucosyltransferase [Medicago truncatula] |
| 6 |
Hb_000417_120 |
0.0941217446 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 7 |
Hb_020648_020 |
0.0991895349 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 8 |
Hb_007765_140 |
0.1046757163 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |
| 9 |
Hb_165404_010 |
0.1049451129 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 10 |
Hb_002276_100 |
0.1099695824 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89A2-like [Jatropha curcas] |
| 11 |
Hb_046101_010 |
0.1103387738 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 12 |
Hb_000075_110 |
0.1107166159 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 13 |
Hb_084127_010 |
0.1115073787 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000907_120 |
0.1115309975 |
transcription factor |
TF Family: G2-like |
PREDICTED: transcription repressor KAN1-like [Populus euphratica] |
| 15 |
Hb_074312_020 |
0.1128548216 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_075449_040 |
0.1155680433 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-1 [Jatropha curcas] |
| 17 |
Hb_020285_010 |
0.1168008402 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3, partial [Pyrus x bretschneideri] |
| 18 |
Hb_000120_070 |
0.1184494716 |
- |
- |
PREDICTED: uncharacterized protein LOC105630351 [Jatropha curcas] |
| 19 |
Hb_002529_080 |
0.1225299671 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase [Jatropha curcas] |
| 20 |
Hb_001369_070 |
0.1227028669 |
- |
- |
unnamed protein product [Vitis vinifera] |