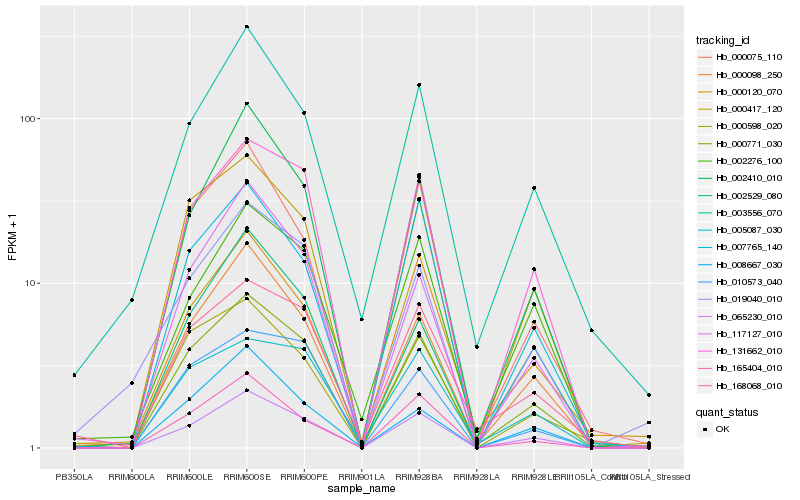
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000771_030 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 2 |
Hb_065230_010 |
0.0636791559 |
- |
- |
PREDICTED: uncharacterized protein LOC100814166 isoform X1 [Glycine max] |
| 3 |
Hb_000417_120 |
0.0734108854 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 4 |
Hb_131662_010 |
0.0779074984 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 5 |
Hb_000098_250 |
0.0885704405 |
- |
- |
UDP-glucose flavonoid 3-O-glucosyltransferase [Medicago truncatula] |
| 6 |
Hb_000598_020 |
0.1036956927 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 7 |
Hb_002410_010 |
0.1041614846 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 8 |
Hb_168068_010 |
0.1094010419 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 9 |
Hb_000075_110 |
0.110233221 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 10 |
Hb_117127_010 |
0.1158724238 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH106 [Jatropha curcas] |
| 11 |
Hb_000120_070 |
0.1171025697 |
- |
- |
PREDICTED: uncharacterized protein LOC105630351 [Jatropha curcas] |
| 12 |
Hb_165404_010 |
0.1209248984 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 13 |
Hb_005087_030 |
0.121357386 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 14 |
Hb_002529_080 |
0.1225195284 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase [Jatropha curcas] |
| 15 |
Hb_010573_040 |
0.1230889978 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal isoform X2 [Populus euphratica] |
| 16 |
Hb_007765_140 |
0.1236812893 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |
| 17 |
Hb_003556_070 |
0.1244158658 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_002276_100 |
0.1287225537 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89A2-like [Jatropha curcas] |
| 19 |
Hb_019040_010 |
0.1319826797 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG7 [Jatropha curcas] |
| 20 |
Hb_008667_030 |
0.1319986741 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |