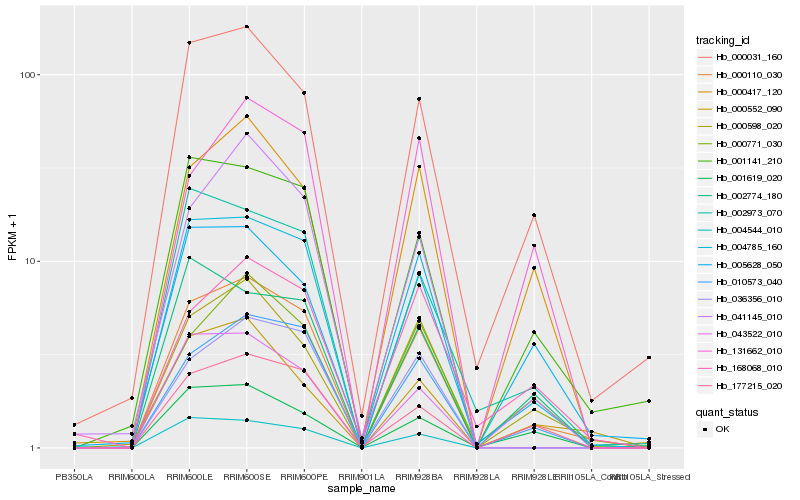
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_030 |
0.0 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_004785_160 |
0.0752510171 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |
| 3 |
Hb_010573_040 |
0.1022614426 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal isoform X2 [Populus euphratica] |
| 4 |
Hb_000598_020 |
0.1083476545 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 5 |
Hb_000031_160 |
0.116076596 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 6 |
Hb_001141_210 |
0.1253382247 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 7 |
Hb_177215_020 |
0.1287498125 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 8 |
Hb_041145_010 |
0.133298287 |
- |
- |
hypothetical protein CISIN_1g021653mg [Citrus sinensis] |
| 9 |
Hb_000417_120 |
0.1348261539 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 10 |
Hb_004544_010 |
0.1360912074 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 11 |
Hb_000771_030 |
0.1363545542 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 12 |
Hb_043522_010 |
0.1408416336 |
- |
- |
- |
| 13 |
Hb_036356_010 |
0.1416902852 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_002973_070 |
0.1455453498 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 15 |
Hb_005628_050 |
0.1505949715 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 16 |
Hb_000552_090 |
0.1515109278 |
- |
- |
hypothetical protein JCGZ_18511 [Jatropha curcas] |
| 17 |
Hb_001619_020 |
0.1555375912 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 18 |
Hb_131662_010 |
0.1598806174 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 19 |
Hb_168068_010 |
0.161523282 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 20 |
Hb_002774_180 |
0.1635618687 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |