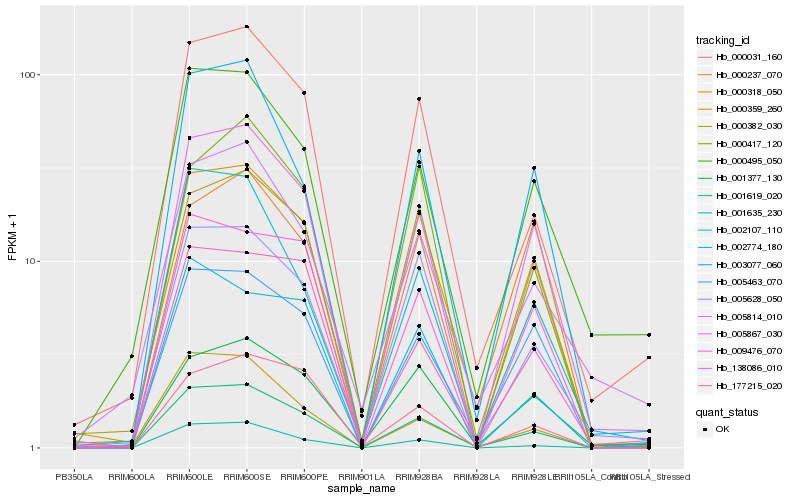
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001619_020 |
0.0 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 2 |
Hb_005867_030 |
0.0856854386 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 3 |
Hb_000031_160 |
0.0907850022 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 4 |
Hb_001635_230 |
0.1072723462 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000237_070 |
0.115701487 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 6 |
Hb_005463_070 |
0.1164446866 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 7 |
Hb_005628_050 |
0.1180520796 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 8 |
Hb_003077_060 |
0.1182425491 |
- |
- |
PREDICTED: 3-oxo-Delta(4,5)-steroid 5-beta-reductase-like [Jatropha curcas] |
| 9 |
Hb_009476_070 |
0.1210160881 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 10 |
Hb_000382_030 |
0.1253395997 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0018s11800g [Populus trichocarpa] |
| 11 |
Hb_000495_050 |
0.1275113541 |
- |
- |
hypothetical protein JCGZ_25174 [Jatropha curcas] |
| 12 |
Hb_177215_020 |
0.1282300598 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 13 |
Hb_002107_110 |
0.1293545055 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000318_050 |
0.1300323043 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002774_180 |
0.1329557376 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 16 |
Hb_005814_010 |
0.1344076584 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_000359_260 |
0.1347285205 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 18 |
Hb_001377_130 |
0.1350455398 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 19 |
Hb_000417_120 |
0.1367409886 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 20 |
Hb_138086_010 |
0.1375482276 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |