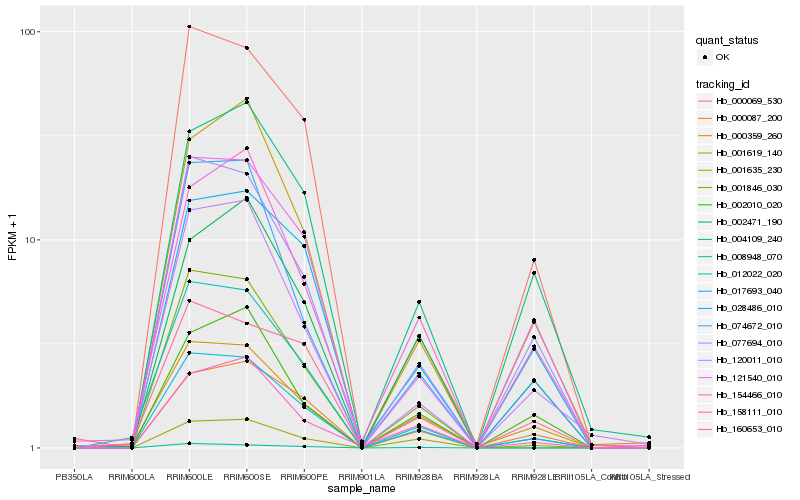
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017693_040 |
0.0 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 2 |
Hb_077694_010 |
0.07477388 |
- |
- |
uncharacterized protein LOC100305624 [Glycine max] |
| 3 |
Hb_121540_010 |
0.0964044123 |
- |
- |
PREDICTED: UDP-glycosyltransferase 88A1-like [Jatropha curcas] |
| 4 |
Hb_000087_200 |
0.0976528059 |
- |
- |
PREDICTED: uncharacterized protein LOC105641954 [Jatropha curcas] |
| 5 |
Hb_001635_230 |
0.1009492987 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000359_260 |
0.1066124746 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 7 |
Hb_012022_020 |
0.1086906106 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0006s10190g [Populus trichocarpa] |
| 8 |
Hb_028486_010 |
0.1092662572 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 9 |
Hb_000069_530 |
0.1124990906 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 10 |
Hb_120011_010 |
0.116044153 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 11 |
Hb_001846_030 |
0.1167490627 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 12 |
Hb_074672_010 |
0.1209252897 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 13 |
Hb_004109_240 |
0.1215629469 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 14 |
Hb_001619_140 |
0.1241197501 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_002471_190 |
0.1243925679 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 16 |
Hb_154466_010 |
0.125630942 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 17 |
Hb_160653_010 |
0.1259434899 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 18 |
Hb_158111_010 |
0.1275370606 |
- |
- |
S-locus-specific glycoprotein precursor, putative [Ricinus communis] |
| 19 |
Hb_002010_020 |
0.1286516071 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 20 |
Hb_008948_070 |
0.1302404859 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |