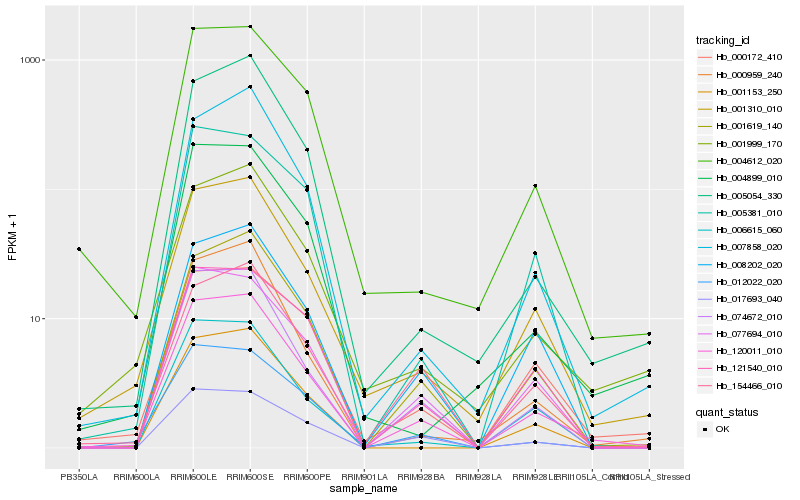
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_120011_010 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 2 |
Hb_074672_010 |
0.0852620389 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 3 |
Hb_001619_140 |
0.0885216069 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 4 |
Hb_001310_010 |
0.0966274117 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type [Jatropha curcas] |
| 5 |
Hb_077694_010 |
0.1036973673 |
- |
- |
uncharacterized protein LOC100305624 [Glycine max] |
| 6 |
Hb_008202_020 |
0.1115061732 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 7 |
Hb_006615_060 |
0.112051089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000959_240 |
0.113655811 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_017693_040 |
0.116044153 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 10 |
Hb_012022_020 |
0.1197633123 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0006s10190g [Populus trichocarpa] |
| 11 |
Hb_004612_020 |
0.1199373149 |
- |
- |
PREDICTED: tetraspanin-8-like [Populus euphratica] |
| 12 |
Hb_005054_330 |
0.1205458139 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 13 |
Hb_001153_250 |
0.121121756 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 1A-like [Jatropha curcas] |
| 14 |
Hb_007858_020 |
0.1211561074 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 15 |
Hb_154466_010 |
0.1228061231 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 16 |
Hb_121540_010 |
0.1242148362 |
- |
- |
PREDICTED: UDP-glycosyltransferase 88A1-like [Jatropha curcas] |
| 17 |
Hb_000172_410 |
0.1247049386 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_005381_010 |
0.1249376216 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001999_170 |
0.1249830079 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 20 |
Hb_004899_010 |
0.1249999075 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |