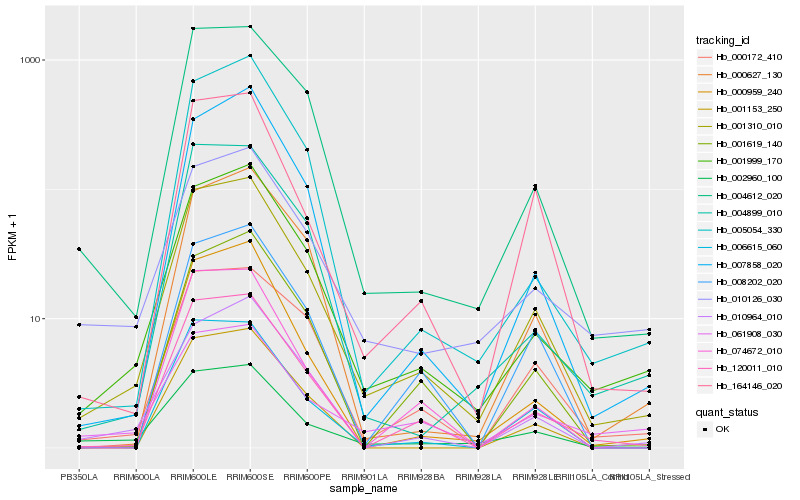
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001310_010 |
0.0 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type [Jatropha curcas] |
| 2 |
Hb_001999_170 |
0.0813485059 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 3 |
Hb_120011_010 |
0.0966274117 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 4 |
Hb_002960_100 |
0.0977978892 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 5 |
Hb_004612_020 |
0.1031546375 |
- |
- |
PREDICTED: tetraspanin-8-like [Populus euphratica] |
| 6 |
Hb_164146_020 |
0.1105671587 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 7 |
Hb_010964_010 |
0.1140336716 |
- |
- |
- |
| 8 |
Hb_006615_060 |
0.1151055824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000172_410 |
0.1166457918 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_007858_020 |
0.1207940994 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 11 |
Hb_000959_240 |
0.1227634678 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_001619_140 |
0.1232673911 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 13 |
Hb_008202_020 |
0.1266368869 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 14 |
Hb_000627_130 |
0.1270472724 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 15 |
Hb_004899_010 |
0.1278190581 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 16 |
Hb_005054_330 |
0.1282161046 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 17 |
Hb_001153_250 |
0.1343280911 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 1A-like [Jatropha curcas] |
| 18 |
Hb_010126_030 |
0.1347567207 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X1 [Jatropha curcas] |
| 19 |
Hb_074672_010 |
0.1399354816 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 20 |
Hb_061908_030 |
0.1416033218 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |