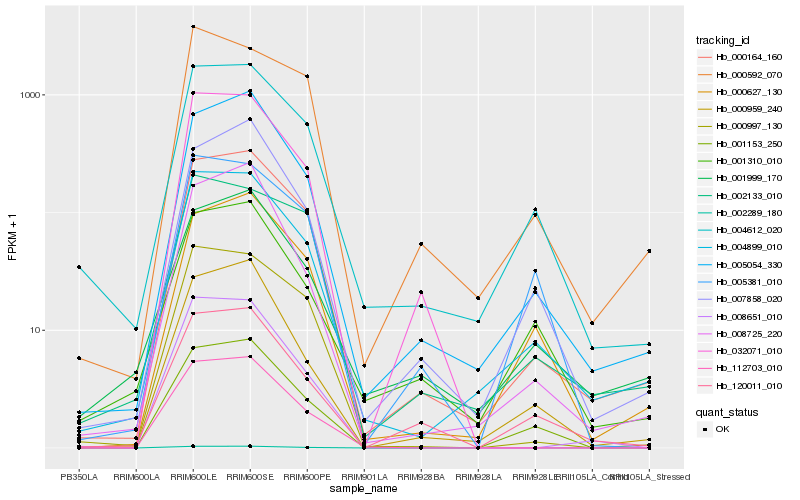
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004899_010 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 2 |
Hb_000164_160 |
0.0807031679 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_004612_020 |
0.0847085361 |
- |
- |
PREDICTED: tetraspanin-8-like [Populus euphratica] |
| 4 |
Hb_005054_330 |
0.106786746 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 5 |
Hb_001153_250 |
0.115365112 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 1A-like [Jatropha curcas] |
| 6 |
Hb_000959_240 |
0.1195246857 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_008651_010 |
0.1207924502 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_000627_130 |
0.1210163745 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 9 |
Hb_000997_130 |
0.12193757 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_112703_010 |
0.1248602574 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Prunus mume] |
| 11 |
Hb_120011_010 |
0.1249999075 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_001310_010 |
0.1278190581 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type [Jatropha curcas] |
| 13 |
Hb_002289_180 |
0.1287967215 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Nelumbo nucifera] |
| 14 |
Hb_001999_170 |
0.1293404118 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 15 |
Hb_000592_070 |
0.1317739952 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 16 |
Hb_002133_010 |
0.1331808587 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 17 |
Hb_005381_010 |
0.1336894816 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 18 |
Hb_008725_220 |
0.1344126427 |
- |
- |
PREDICTED: uncharacterized protein LOC105642593 [Jatropha curcas] |
| 19 |
Hb_032071_010 |
0.1354017372 |
- |
- |
hypothetical protein EUGRSUZ_J02969 [Eucalyptus grandis] |
| 20 |
Hb_007858_020 |
0.1360584073 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |