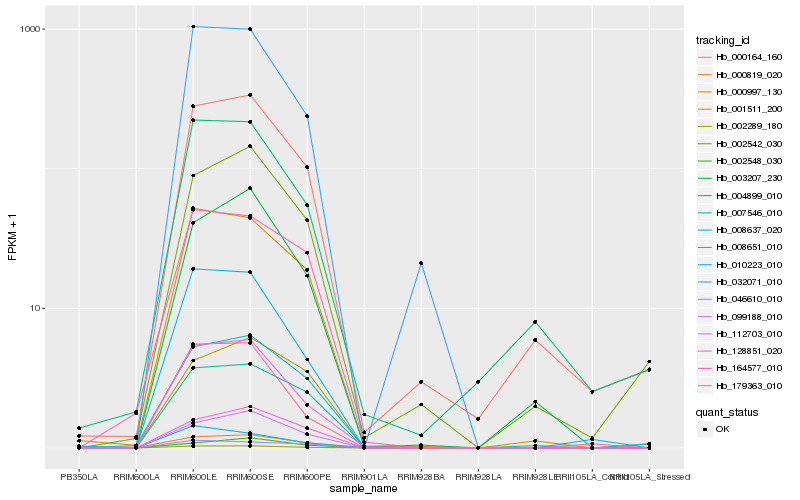
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002289_180 |
0.0 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Nelumbo nucifera] |
| 2 |
Hb_112703_010 |
0.0555435996 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Prunus mume] |
| 3 |
Hb_000997_130 |
0.0696621251 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_032071_010 |
0.0795682756 |
- |
- |
hypothetical protein EUGRSUZ_J02969 [Eucalyptus grandis] |
| 5 |
Hb_008651_010 |
0.0850781508 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_099188_010 |
0.0906192248 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 7 |
Hb_008637_020 |
0.0933142907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000164_160 |
0.095548922 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_179363_010 |
0.0957820278 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 10 |
Hb_007546_010 |
0.1029365473 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_164577_010 |
0.106518824 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 12 |
Hb_128851_020 |
0.1121825265 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_046610_010 |
0.115089748 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_010223_010 |
0.1166209162 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 15 |
Hb_003207_230 |
0.1205602956 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 16 |
Hb_000819_020 |
0.1234223763 |
- |
- |
hypothetical protein PRUPE_ppa020290mg, partial [Prunus persica] |
| 17 |
Hb_004899_010 |
0.1287967215 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 18 |
Hb_002542_030 |
0.1316075578 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 19 |
Hb_001511_200 |
0.1323749567 |
- |
- |
Embryonic abundant protein, putative [Ricinus communis] |
| 20 |
Hb_002548_030 |
0.1327904457 |
- |
- |
PREDICTED: uncharacterized protein LOC103343650 [Prunus mume] |