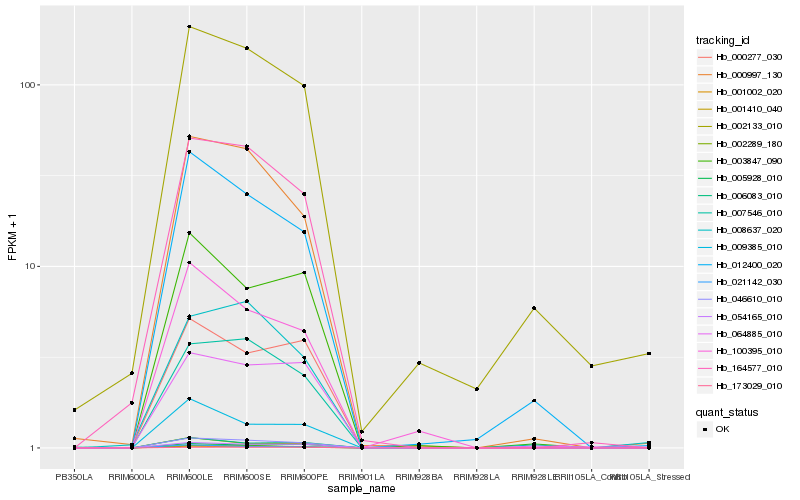
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_046610_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_054165_010 |
0.0688439265 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 3 |
Hb_164577_010 |
0.0714210836 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 4 |
Hb_000997_130 |
0.0800126133 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_001002_020 |
0.085247782 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_007546_010 |
0.0890098096 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_021142_030 |
0.1032310337 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 8 |
Hb_012400_020 |
0.103956108 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 9 |
Hb_006083_010 |
0.1041325622 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_000277_030 |
0.1065879256 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 11 |
Hb_005928_010 |
0.1093576413 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_001410_040 |
0.1144409651 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |
| 13 |
Hb_009385_010 |
0.1148443304 |
- |
- |
Cysteine/Histidine-rich C1 domain family protein, putative [Theobroma cacao] |
| 14 |
Hb_002289_180 |
0.115089748 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Nelumbo nucifera] |
| 15 |
Hb_100395_010 |
0.1157439694 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_003847_090 |
0.117315422 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |
| 17 |
Hb_064885_010 |
0.1207567632 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 18 |
Hb_008637_020 |
0.1226056732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002133_010 |
0.1240493768 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 20 |
Hb_173029_010 |
0.1273544333 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |