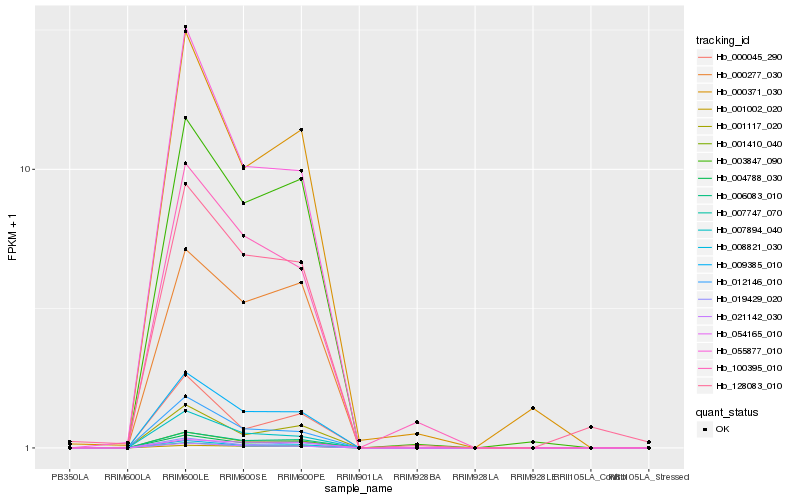
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001410_040 |
0.0 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |
| 2 |
Hb_009385_010 |
0.0094171995 |
- |
- |
Cysteine/Histidine-rich C1 domain family protein, putative [Theobroma cacao] |
| 3 |
Hb_004788_030 |
0.0226557548 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_008821_030 |
0.0320248824 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 5 |
Hb_021142_030 |
0.0334231119 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 6 |
Hb_006083_010 |
0.033690222 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_007747_070 |
0.0381088743 |
- |
- |
PREDICTED: casparian strip membrane protein 1-like [Jatropha curcas] |
| 8 |
Hb_003847_090 |
0.0662989812 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |
| 9 |
Hb_007894_040 |
0.067873061 |
- |
- |
PREDICTED: NEP1-interacting protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_019429_020 |
0.0703876741 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_012146_010 |
0.0752854268 |
- |
- |
PREDICTED: uncharacterized protein LOC105111251 [Populus euphratica] |
| 12 |
Hb_001117_020 |
0.0792861786 |
- |
- |
hypothetical protein CICLE_v10008579mg [Citrus clementina] |
| 13 |
Hb_055877_010 |
0.0797269826 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 14 |
Hb_000371_030 |
0.0929412843 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: anthocyanidin 3-O-glucosyltransferase 5 [Prunus mume] |
| 15 |
Hb_000277_030 |
0.0947259295 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 16 |
Hb_100395_010 |
0.0984307447 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_054165_010 |
0.1015440879 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 18 |
Hb_001002_020 |
0.1074877082 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_000045_290 |
0.1093567984 |
- |
- |
- |
| 20 |
Hb_128083_010 |
0.1114198487 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |