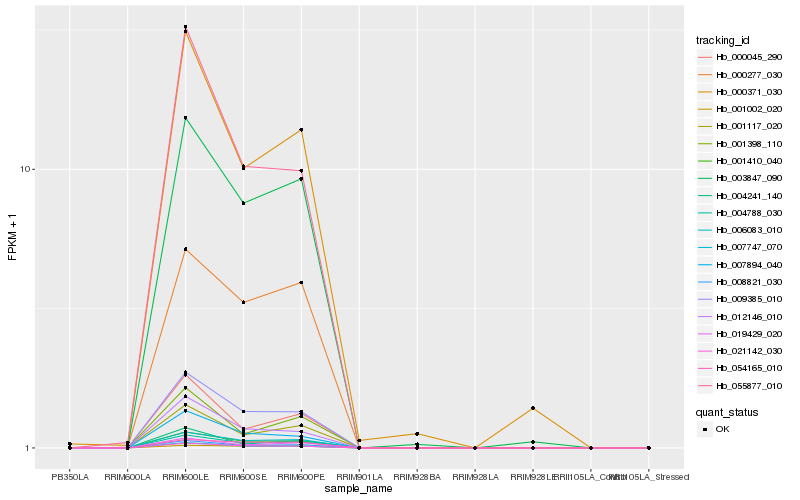
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004788_030 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_008821_030 |
0.0177433168 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 3 |
Hb_001410_040 |
0.0226557548 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |
| 4 |
Hb_009385_010 |
0.0301039514 |
- |
- |
Cysteine/Histidine-rich C1 domain family protein, putative [Theobroma cacao] |
| 5 |
Hb_006083_010 |
0.0337396056 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_019429_020 |
0.0478406953 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_007747_070 |
0.0508451859 |
- |
- |
PREDICTED: casparian strip membrane protein 1-like [Jatropha curcas] |
| 8 |
Hb_021142_030 |
0.0554915491 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 9 |
Hb_001117_020 |
0.0580514882 |
- |
- |
hypothetical protein CICLE_v10008579mg [Citrus clementina] |
| 10 |
Hb_003847_090 |
0.0610842861 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |
| 11 |
Hb_007894_040 |
0.0822683863 |
- |
- |
PREDICTED: NEP1-interacting protein 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_055877_010 |
0.0845811993 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 13 |
Hb_000371_030 |
0.084734382 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: anthocyanidin 3-O-glucosyltransferase 5 [Prunus mume] |
| 14 |
Hb_012146_010 |
0.0852809439 |
- |
- |
PREDICTED: uncharacterized protein LOC105111251 [Populus euphratica] |
| 15 |
Hb_000277_030 |
0.0913682621 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 16 |
Hb_000045_290 |
0.0930601718 |
- |
- |
- |
| 17 |
Hb_001398_110 |
0.1045839629 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 18 |
Hb_054165_010 |
0.106940372 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 19 |
Hb_004241_140 |
0.1087319457 |
- |
- |
S-locus lectin protein kinase family protein, putative [Theobroma cacao] |
| 20 |
Hb_001002_020 |
0.1095348842 |
- |
- |
ATP binding protein, putative [Ricinus communis] |