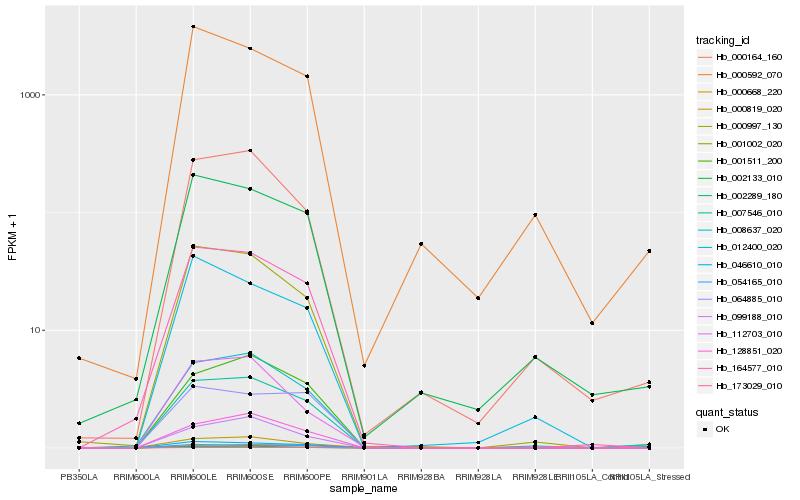
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164577_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 2 |
Hb_046610_010 |
0.0714210836 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_000997_130 |
0.0817818342 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_007546_010 |
0.0936125743 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_008637_020 |
0.0936681494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002289_180 |
0.106518824 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Nelumbo nucifera] |
| 7 |
Hb_000819_020 |
0.1130504132 |
- |
- |
hypothetical protein PRUPE_ppa020290mg, partial [Prunus persica] |
| 8 |
Hb_054165_010 |
0.1153215994 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 9 |
Hb_002133_010 |
0.1179012394 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 10 |
Hb_001002_020 |
0.1285350991 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_001511_200 |
0.1286198662 |
- |
- |
Embryonic abundant protein, putative [Ricinus communis] |
| 12 |
Hb_128851_020 |
0.1319832946 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_012400_020 |
0.1332558647 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 14 |
Hb_064885_010 |
0.1333072118 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 15 |
Hb_000164_160 |
0.1334961179 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000592_070 |
0.1419008005 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 17 |
Hb_099188_010 |
0.1428371749 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 18 |
Hb_000668_220 |
0.144104072 |
- |
- |
PREDICTED: uncharacterized protein LOC105119281 [Populus euphratica] |
| 19 |
Hb_112703_010 |
0.1444276494 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Prunus mume] |
| 20 |
Hb_173029_010 |
0.144973764 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |