| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
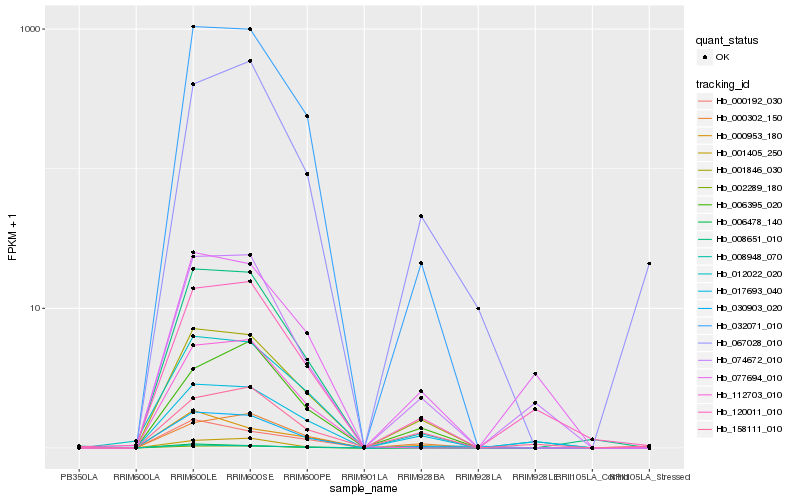
Hb_001846_030 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 2 |
Hb_008948_070 |
0.0811930283 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_032071_010 |
0.0973031128 |
- |
- |
hypothetical protein EUGRSUZ_J02969 [Eucalyptus grandis] |
| 4 |
Hb_006478_140 |
0.1117275208 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |
| 5 |
Hb_012022_020 |
0.1125775863 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0006s10190g [Populus trichocarpa] |
| 6 |
Hb_017693_040 |
0.1167490627 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 7 |
Hb_000192_030 |
0.1304910097 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 8 |
Hb_001405_250 |
0.1333900235 |
- |
- |
PREDICTED: uncharacterized protein LOC105645451 [Jatropha curcas] |
| 9 |
Hb_074672_010 |
0.1375810651 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 10 |
Hb_000953_180 |
0.1390519973 |
- |
- |
- |
| 11 |
Hb_006395_020 |
0.1397377484 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 12 |
Hb_000302_150 |
0.1434899197 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1581940 [Ricinus communis] |
| 13 |
Hb_030903_020 |
0.1456065937 |
- |
- |
PREDICTED: putative 12-oxophytodienoate reductase 11 [Jatropha curcas] |
| 14 |
Hb_067028_010 |
0.1544163584 |
- |
- |
BnaC03g72040D [Brassica napus] |
| 15 |
Hb_077694_010 |
0.1546194092 |
- |
- |
uncharacterized protein LOC100305624 [Glycine max] |
| 16 |
Hb_120011_010 |
0.1548426988 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 17 |
Hb_002289_180 |
0.1549873194 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Nelumbo nucifera] |
| 18 |
Hb_112703_010 |
0.1589057725 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Prunus mume] |
| 19 |
Hb_008651_010 |
0.1601642177 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_158111_010 |
0.1603269888 |
- |
- |
S-locus-specific glycoprotein precursor, putative [Ricinus communis] |