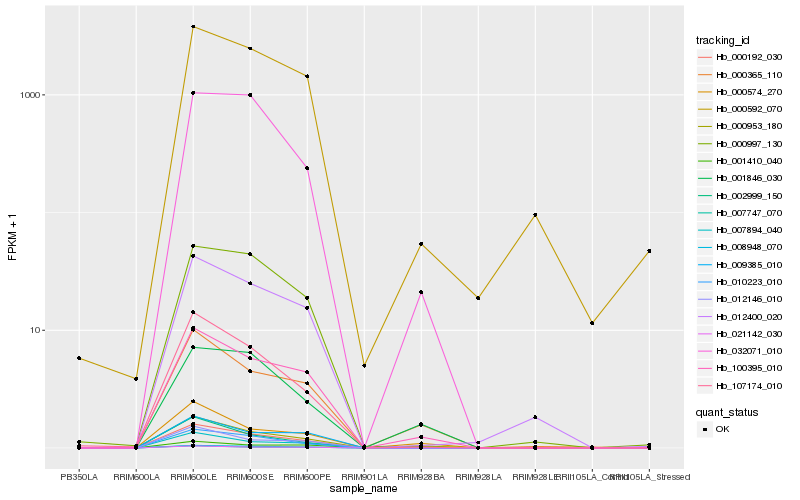
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000192_030 |
0.0 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 2 |
Hb_000953_180 |
0.0539521429 |
- |
- |
- |
| 3 |
Hb_100395_010 |
0.058545562 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_010223_010 |
0.0992631426 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 5 |
Hb_000365_110 |
0.0994537125 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 6 |
Hb_008948_070 |
0.1018659968 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_107174_010 |
0.1067203849 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_021142_030 |
0.112498611 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 9 |
Hb_007894_040 |
0.1128352414 |
- |
- |
PREDICTED: NEP1-interacting protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_012400_020 |
0.1165366933 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 11 |
Hb_000574_270 |
0.1196728763 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 12 |
Hb_007747_070 |
0.1245780241 |
- |
- |
PREDICTED: casparian strip membrane protein 1-like [Jatropha curcas] |
| 13 |
Hb_012146_010 |
0.1250577289 |
- |
- |
PREDICTED: uncharacterized protein LOC105111251 [Populus euphratica] |
| 14 |
Hb_032071_010 |
0.1260853827 |
- |
- |
hypothetical protein EUGRSUZ_J02969 [Eucalyptus grandis] |
| 15 |
Hb_000592_070 |
0.1280097249 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 16 |
Hb_000997_130 |
0.1285644275 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_001846_030 |
0.1304910097 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 18 |
Hb_009385_010 |
0.1307616016 |
- |
- |
Cysteine/Histidine-rich C1 domain family protein, putative [Theobroma cacao] |
| 19 |
Hb_002999_150 |
0.1327770149 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 20 |
Hb_001410_040 |
0.1369162901 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |