| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
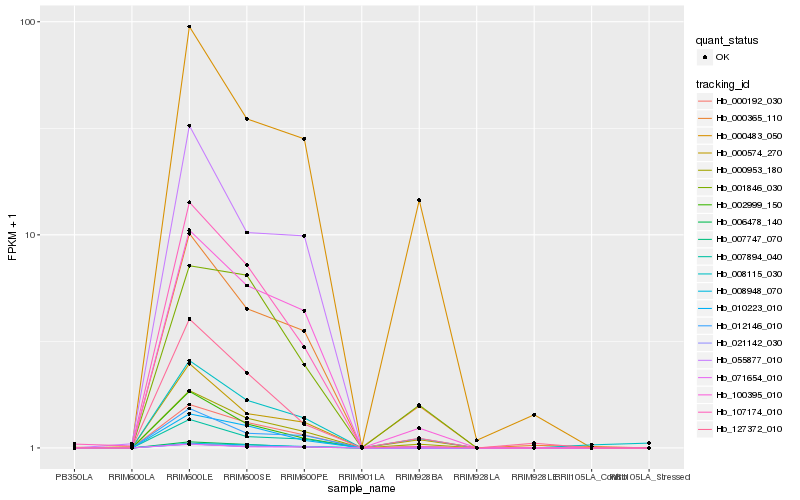
Hb_000953_180 |
0.0 |
- |
- |
- |
| 2 |
Hb_000192_030 |
0.0539521429 |
- |
- |
PREDICTED: TATA-box-binding protein 2 isoform X1 [Setaria italica] |
| 3 |
Hb_000365_110 |
0.0652365251 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 4 |
Hb_000574_270 |
0.0711156581 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 5 |
Hb_100395_010 |
0.0816347195 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_008948_070 |
0.0923832435 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000483_050 |
0.1219745626 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 8 |
Hb_006478_140 |
0.1238708957 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |
| 9 |
Hb_127372_010 |
0.126403948 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |
| 10 |
Hb_008115_030 |
0.1287578818 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 11 |
Hb_107174_010 |
0.1300316395 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_007894_040 |
0.1304766243 |
- |
- |
PREDICTED: NEP1-interacting protein 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012146_010 |
0.1368037586 |
- |
- |
PREDICTED: uncharacterized protein LOC105111251 [Populus euphratica] |
| 14 |
Hb_010223_010 |
0.1389626748 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 15 |
Hb_001846_030 |
0.1390519973 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 16 |
Hb_021142_030 |
0.1401174898 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 17 |
Hb_002999_150 |
0.1412056677 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 18 |
Hb_007747_070 |
0.1427876667 |
- |
- |
PREDICTED: casparian strip membrane protein 1-like [Jatropha curcas] |
| 19 |
Hb_071654_010 |
0.1467607957 |
- |
- |
non-ltr retroelement reverse transcriptase [Rosa rugosa] |
| 20 |
Hb_055877_010 |
0.148348761 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |