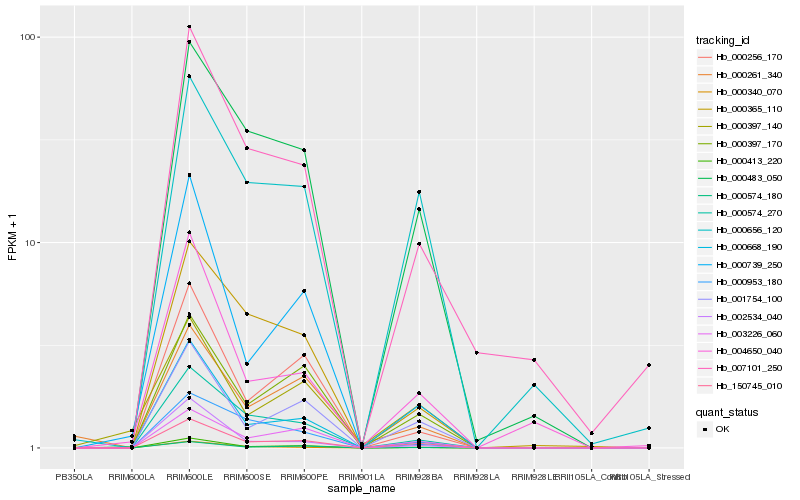
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_150745_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |
| 2 |
Hb_001754_100 |
0.0906834626 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 3 |
Hb_000413_220 |
0.0961641881 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 4 |
Hb_000574_180 |
0.1059588519 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 5 |
Hb_000483_050 |
0.1122151679 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 6 |
Hb_000397_170 |
0.1125965467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000574_270 |
0.1150906573 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 8 |
Hb_003226_060 |
0.1174464795 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 9 |
Hb_000656_120 |
0.1284319378 |
transcription factor |
TF Family: Tify |
JAZ2 [Hevea brasiliensis] |
| 10 |
Hb_000340_070 |
0.1321744201 |
- |
- |
PREDICTED: CSC1-like protein At4g02900 [Jatropha curcas] |
| 11 |
Hb_000668_190 |
0.1397046 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 12 |
Hb_007101_250 |
0.1457096512 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 13 |
Hb_000256_170 |
0.1498096219 |
- |
- |
PREDICTED: uncharacterized protein LOC105638320 [Jatropha curcas] |
| 14 |
Hb_000365_110 |
0.1523652919 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 15 |
Hb_004650_040 |
0.1614320892 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 16 |
Hb_002534_040 |
0.1624511031 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 17 |
Hb_000739_250 |
0.1685334464 |
- |
- |
PREDICTED: uncharacterized protein LOC105120821 [Populus euphratica] |
| 18 |
Hb_000397_140 |
0.1758910663 |
transcription factor |
TF Family: PLATZ |
hypothetical protein JCGZ_16980 [Jatropha curcas] |
| 19 |
Hb_000261_340 |
0.1782537147 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 20 |
Hb_000953_180 |
0.1800336778 |
- |
- |
- |