| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
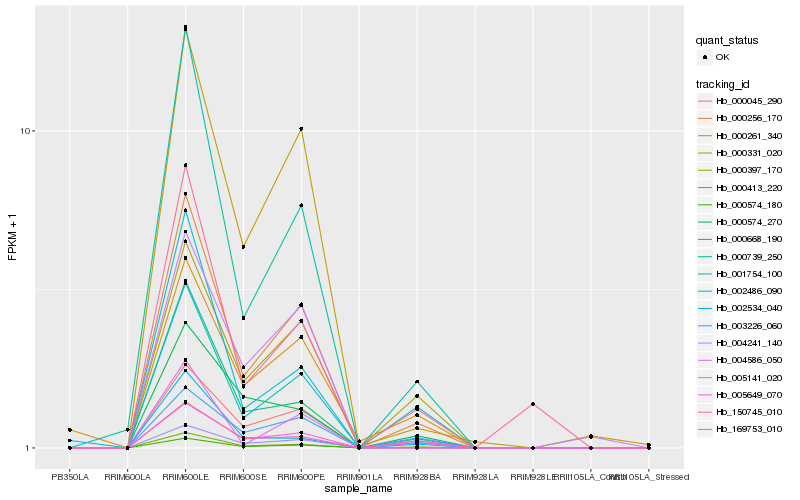
Hb_000574_180 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 2 |
Hb_000413_220 |
0.0411329355 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 3 |
Hb_000256_170 |
0.0464185818 |
- |
- |
PREDICTED: uncharacterized protein LOC105638320 [Jatropha curcas] |
| 4 |
Hb_000739_250 |
0.0925801382 |
- |
- |
PREDICTED: uncharacterized protein LOC105120821 [Populus euphratica] |
| 5 |
Hb_000668_190 |
0.095253605 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 6 |
Hb_000397_170 |
0.0984786856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001754_100 |
0.1026575839 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 8 |
Hb_150745_010 |
0.1059588519 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |
| 9 |
Hb_003226_060 |
0.1115413733 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 10 |
Hb_000574_270 |
0.1326946726 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 11 |
Hb_000331_020 |
0.1425901918 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_005649_070 |
0.1432425237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004241_140 |
0.1474454558 |
- |
- |
S-locus lectin protein kinase family protein, putative [Theobroma cacao] |
| 14 |
Hb_005141_020 |
0.1475647336 |
- |
- |
laccase, putative [Ricinus communis] |
| 15 |
Hb_004586_050 |
0.1522396014 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 16 |
Hb_000261_340 |
0.1546573871 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 17 |
Hb_002486_090 |
0.1549727213 |
- |
- |
hypothetical protein POPTR_0005s06910g [Populus trichocarpa] |
| 18 |
Hb_002534_040 |
0.1550000011 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 19 |
Hb_000045_290 |
0.155685825 |
- |
- |
- |
| 20 |
Hb_169753_010 |
0.1568731337 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |