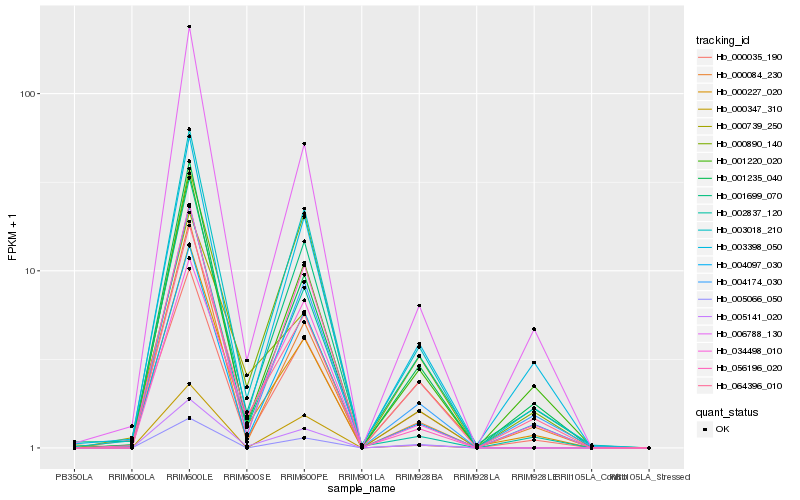
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005141_020 |
0.0 |
- |
- |
laccase, putative [Ricinus communis] |
| 2 |
Hb_003018_210 |
0.0756031586 |
- |
- |
PREDICTED: uncharacterized protein LOC105138721 isoform X2 [Populus euphratica] |
| 3 |
Hb_000035_190 |
0.1027493661 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002837_120 |
0.1031442053 |
- |
- |
PREDICTED: legumin A-like [Jatropha curcas] |
| 5 |
Hb_034498_010 |
0.1053154475 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 6 |
Hb_004174_030 |
0.1054216399 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Populus euphratica] |
| 7 |
Hb_000347_310 |
0.1077760098 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 3-like [Jatropha curcas] |
| 8 |
Hb_004097_030 |
0.1160912061 |
- |
- |
PREDICTED: uncharacterized protein LOC105634518 [Jatropha curcas] |
| 9 |
Hb_001699_070 |
0.1169421457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_064396_010 |
0.1175180158 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000739_250 |
0.1181471708 |
- |
- |
PREDICTED: uncharacterized protein LOC105120821 [Populus euphratica] |
| 12 |
Hb_006788_130 |
0.1189461413 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 13 |
Hb_005066_050 |
0.1205763387 |
- |
- |
- |
| 14 |
Hb_001235_040 |
0.1222789204 |
- |
- |
hypothetical protein POPTR_0001s47470g [Populus trichocarpa] |
| 15 |
Hb_001220_020 |
0.1236984822 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase IMK2 [Jatropha curcas] |
| 16 |
Hb_000084_230 |
0.1237936474 |
- |
- |
hypothetical protein POPTR_0016s08030g [Populus trichocarpa] |
| 17 |
Hb_000227_020 |
0.1248264292 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 18 |
Hb_000890_140 |
0.1253460905 |
- |
- |
PREDICTED: uncharacterized protein LOC105646185 [Jatropha curcas] |
| 19 |
Hb_056196_020 |
0.1276024191 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003398_050 |
0.1277438791 |
- |
- |
B-type cell cycle switch protein ccs52B [Populus trichocarpa] |