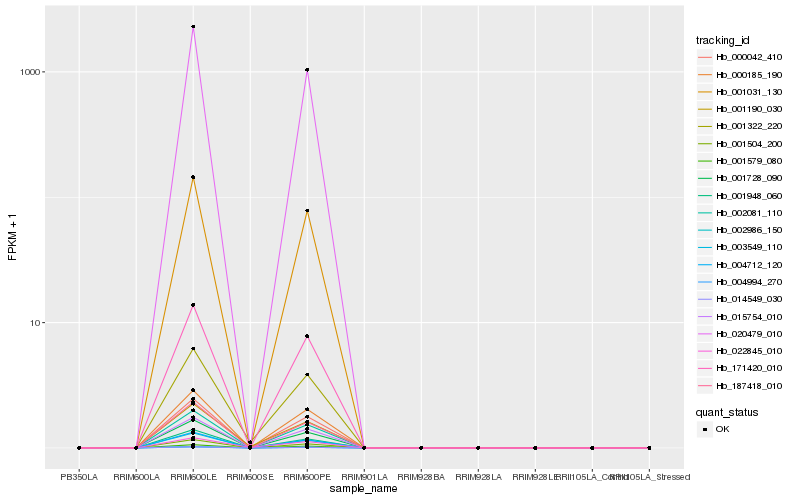
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001190_030 |
0.0 |
- |
- |
hypothetical protein VITISV_017115 [Vitis vinifera] |
| 2 |
Hb_001322_220 |
0.0190556028 |
- |
- |
Major pollen allergen Ory s 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_022845_010 |
0.0613818068 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 4 |
Hb_002986_150 |
0.0850458779 |
- |
- |
hypothetical protein glysoja_022060 [Glycine soja] |
| 5 |
Hb_001728_090 |
0.0850533 |
transcription factor |
TF Family: MYB |
MYB family protein [Jatropha curcas] |
| 6 |
Hb_003549_110 |
0.085555011 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 7 |
Hb_004712_120 |
0.0855761788 |
- |
- |
Pectinesterase U1 precursor, putative [Ricinus communis] |
| 8 |
Hb_004994_270 |
0.0856564022 |
- |
- |
PREDICTED: tRNA-specific adenosine deaminase 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_014549_030 |
0.085759458 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 10 |
Hb_000042_410 |
0.0858279109 |
- |
- |
Patatin precursor, putative [Ricinus communis] |
| 11 |
Hb_001504_200 |
0.0858664913 |
- |
- |
Beta-3 adrenergic receptor, putative [Theobroma cacao] |
| 12 |
Hb_171420_010 |
0.0859547276 |
- |
- |
hypothetical protein AMTR_s00126p00056890 [Amborella trichopoda] |
| 13 |
Hb_002081_110 |
0.0863586974 |
- |
- |
PREDICTED: uncharacterized protein LOC105129793 isoform X1 [Populus euphratica] |
| 14 |
Hb_001031_130 |
0.0863745596 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_187418_010 |
0.0866305117 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Citrus sinensis] |
| 16 |
Hb_020479_010 |
0.0871287231 |
- |
- |
cinnamyl alcohol dehydrogenase [Pyrus pyrifolia] |
| 17 |
Hb_000185_190 |
0.0871563168 |
- |
- |
hypothetical protein L484_017526 [Morus notabilis] |
| 18 |
Hb_001948_060 |
0.087406043 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_015754_010 |
0.0875031714 |
- |
- |
Galactose oxidase precursor, putative [Ricinus communis] |
| 20 |
Hb_001579_080 |
0.087999701 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |