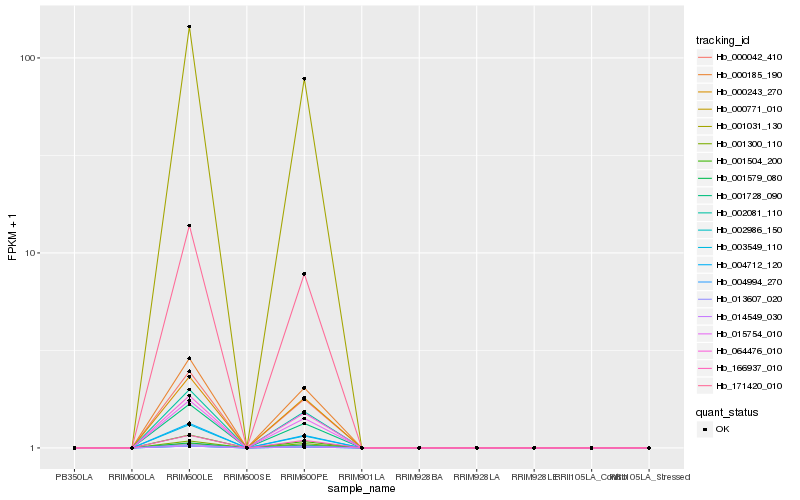
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001031_130 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_002081_110 |
9.1221e-05 |
- |
- |
PREDICTED: uncharacterized protein LOC105129793 isoform X1 [Populus euphratica] |
| 3 |
Hb_171420_010 |
0.0026291999 |
- |
- |
hypothetical protein AMTR_s00126p00056890 [Amborella trichopoda] |
| 4 |
Hb_000042_410 |
0.0035366026 |
- |
- |
Patatin precursor, putative [Ricinus communis] |
| 5 |
Hb_000185_190 |
0.0039809871 |
- |
- |
hypothetical protein L484_017526 [Morus notabilis] |
| 6 |
Hb_014549_030 |
0.0040565177 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 7 |
Hb_004994_270 |
0.0048878129 |
- |
- |
PREDICTED: tRNA-specific adenosine deaminase 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_015754_010 |
0.0055128948 |
- |
- |
Galactose oxidase precursor, putative [Ricinus communis] |
| 9 |
Hb_001579_080 |
0.0075360178 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 10 |
Hb_000771_010 |
0.0114688881 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_002986_150 |
0.0142479614 |
- |
- |
hypothetical protein glysoja_022060 [Glycine soja] |
| 12 |
Hb_166937_010 |
0.0156167214 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_001728_090 |
0.0166353764 |
transcription factor |
TF Family: MYB |
MYB family protein [Jatropha curcas] |
| 14 |
Hb_013607_020 |
0.0172178879 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_064476_010 |
0.0212160923 |
- |
- |
PREDICTED: beta-Amyrin Synthase 1-like [Sesamum indicum] |
| 16 |
Hb_003549_110 |
0.024573413 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 17 |
Hb_004712_120 |
0.0247655012 |
- |
- |
Pectinesterase U1 precursor, putative [Ricinus communis] |
| 18 |
Hb_000243_270 |
0.0252378441 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 19 |
Hb_001300_110 |
0.0262684639 |
- |
- |
PREDICTED: bidirectional sugar transporter N3-like [Jatropha curcas] |
| 20 |
Hb_001504_200 |
0.0270952507 |
- |
- |
Beta-3 adrenergic receptor, putative [Theobroma cacao] |