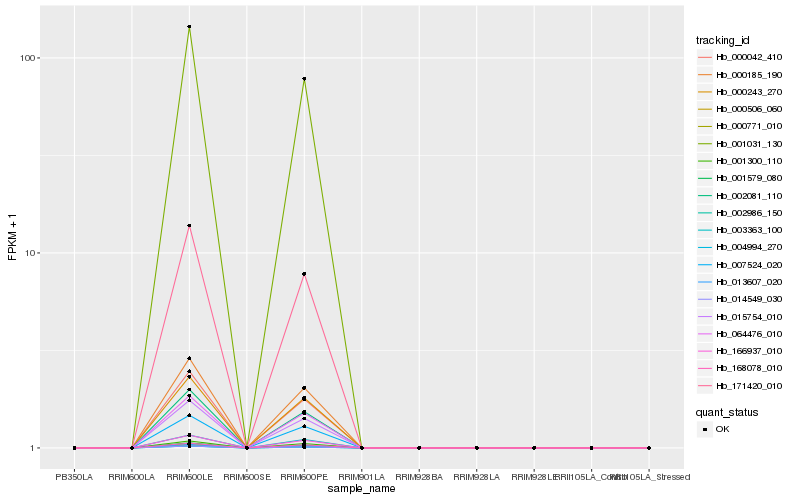
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000771_010 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_001579_080 |
0.0039330092 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_166937_010 |
0.0041481346 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_013607_020 |
0.0057494596 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_015754_010 |
0.0059561474 |
- |
- |
Galactose oxidase precursor, putative [Ricinus communis] |
| 6 |
Hb_000185_190 |
0.0074880411 |
- |
- |
hypothetical protein L484_017526 [Morus notabilis] |
| 7 |
Hb_064476_010 |
0.0097481618 |
- |
- |
PREDICTED: beta-Amyrin Synthase 1-like [Sesamum indicum] |
| 8 |
Hb_001031_130 |
0.0114688881 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_002081_110 |
0.0115601042 |
- |
- |
PREDICTED: uncharacterized protein LOC105129793 isoform X1 [Populus euphratica] |
| 10 |
Hb_000243_270 |
0.0137705597 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 11 |
Hb_171420_010 |
0.0140979128 |
- |
- |
hypothetical protein AMTR_s00126p00056890 [Amborella trichopoda] |
| 12 |
Hb_001300_110 |
0.0148013684 |
- |
- |
PREDICTED: bidirectional sugar transporter N3-like [Jatropha curcas] |
| 13 |
Hb_000042_410 |
0.0150052397 |
- |
- |
Patatin precursor, putative [Ricinus communis] |
| 14 |
Hb_014549_030 |
0.0155251078 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 15 |
Hb_004994_270 |
0.0163563224 |
- |
- |
PREDICTED: tRNA-specific adenosine deaminase 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007524_020 |
0.0170201929 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA4 [Jatropha curcas] |
| 17 |
Hb_000506_060 |
0.0206229759 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease [Malus domestica] |
| 18 |
Hb_003363_100 |
0.0229835289 |
- |
- |
- |
| 19 |
Hb_002986_150 |
0.0257150996 |
- |
- |
hypothetical protein glysoja_022060 [Glycine soja] |
| 20 |
Hb_168078_010 |
0.026011077 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like, partial [Jatropha curcas] |