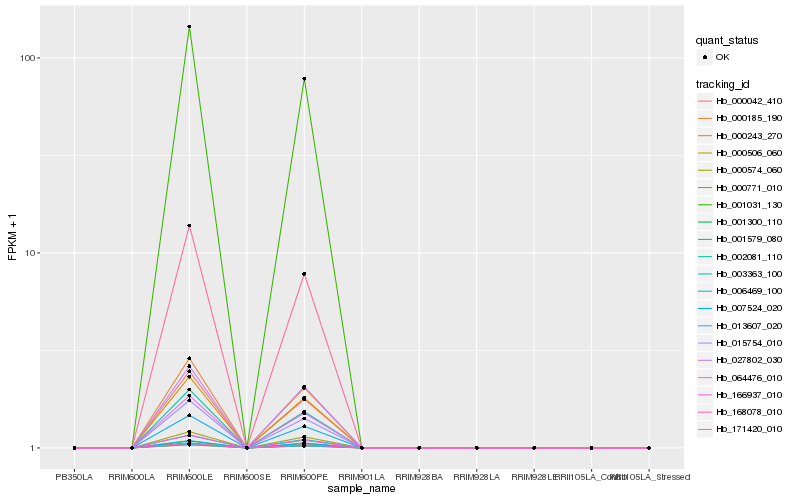
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_064476_010 |
0.0 |
- |
- |
PREDICTED: beta-Amyrin Synthase 1-like [Sesamum indicum] |
| 2 |
Hb_013607_020 |
0.0039987914 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000243_270 |
0.0040226115 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 4 |
Hb_001300_110 |
0.0050534948 |
- |
- |
PREDICTED: bidirectional sugar transporter N3-like [Jatropha curcas] |
| 5 |
Hb_166937_010 |
0.0056001175 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_007524_020 |
0.0072725071 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA4 [Jatropha curcas] |
| 7 |
Hb_000771_010 |
0.0097481618 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_000506_060 |
0.0108756743 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease [Malus domestica] |
| 9 |
Hb_003363_100 |
0.0132365318 |
- |
- |
- |
| 10 |
Hb_001579_080 |
0.0136809606 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 11 |
Hb_015754_010 |
0.0157039428 |
- |
- |
Galactose oxidase precursor, putative [Ricinus communis] |
| 12 |
Hb_168078_010 |
0.0162645311 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like, partial [Jatropha curcas] |
| 13 |
Hb_000185_190 |
0.0172356966 |
- |
- |
hypothetical protein L484_017526 [Morus notabilis] |
| 14 |
Hb_027802_030 |
0.0189568805 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0017s11800g [Populus trichocarpa] |
| 15 |
Hb_000574_060 |
0.0200197905 |
- |
- |
hypothetical protein F775_19731 [Aegilops tauschii] |
| 16 |
Hb_001031_130 |
0.0212160923 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_002081_110 |
0.0213072966 |
- |
- |
PREDICTED: uncharacterized protein LOC105129793 isoform X1 [Populus euphratica] |
| 18 |
Hb_006469_100 |
0.0215648119 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 19 |
Hb_171420_010 |
0.0238447487 |
- |
- |
hypothetical protein AMTR_s00126p00056890 [Amborella trichopoda] |
| 20 |
Hb_000042_410 |
0.0247519355 |
- |
- |
Patatin precursor, putative [Ricinus communis] |