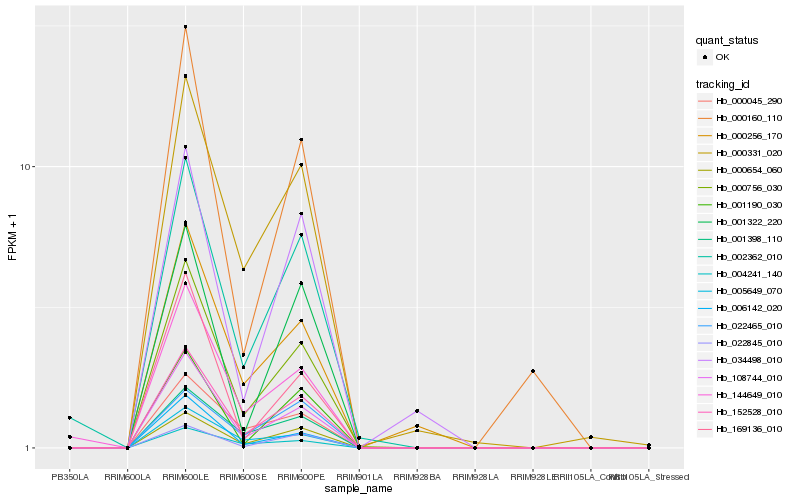
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_152528_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 2 |
Hb_000756_030 |
0.0370862144 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_108744_010 |
0.0393950068 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 4 |
Hb_000654_060 |
0.048278452 |
- |
- |
hypothetical protein CICLE_v10027550mg [Citrus clementina] |
| 5 |
Hb_001398_110 |
0.087303284 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 6 |
Hb_022845_010 |
0.0935558809 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 7 |
Hb_004241_140 |
0.0966664945 |
- |
- |
S-locus lectin protein kinase family protein, putative [Theobroma cacao] |
| 8 |
Hb_000045_290 |
0.0998151736 |
- |
- |
- |
| 9 |
Hb_000331_020 |
0.1004639798 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_006142_020 |
0.1010521578 |
- |
- |
JHL25P11.12 [Jatropha curcas] |
| 11 |
Hb_005649_070 |
0.1039991606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001190_030 |
0.1096188245 |
- |
- |
hypothetical protein VITISV_017115 [Vitis vinifera] |
| 13 |
Hb_002362_010 |
0.1126570526 |
- |
- |
hypothetical protein JCGZ_12328 [Jatropha curcas] |
| 14 |
Hb_001322_220 |
0.1203236978 |
- |
- |
Major pollen allergen Ory s 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_144649_010 |
0.1210561574 |
- |
- |
60S ribosomal protein L34A [Hevea brasiliensis] |
| 16 |
Hb_000160_110 |
0.1219572282 |
- |
- |
PREDICTED: uncharacterized protein LOC100253584 [Vitis vinifera] |
| 17 |
Hb_000256_170 |
0.12492213 |
- |
- |
PREDICTED: uncharacterized protein LOC105638320 [Jatropha curcas] |
| 18 |
Hb_169136_010 |
0.1268739876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_034498_010 |
0.1312808263 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 20 |
Hb_022465_010 |
0.1316097328 |
- |
- |
hypothetical protein JCGZ_06012 [Jatropha curcas] |