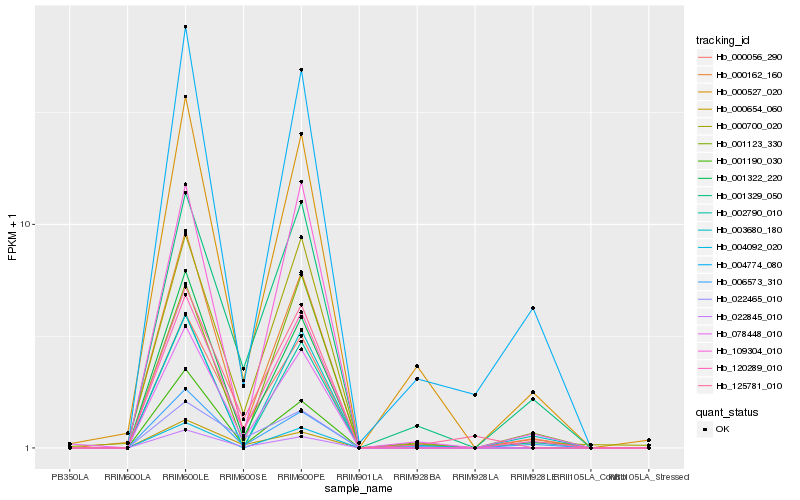
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_120289_010 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 2 |
Hb_000056_290 |
0.0703754934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000162_160 |
0.0896633623 |
- |
- |
PREDICTED: auxin transporter-like protein 5 [Populus euphratica] |
| 4 |
Hb_022845_010 |
0.0898520512 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 5 |
Hb_000700_020 |
0.0918279783 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 6 |
Hb_001329_050 |
0.1012597229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001322_220 |
0.1115326656 |
- |
- |
Major pollen allergen Ory s 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_109304_010 |
0.1201782953 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 9 |
Hb_006573_310 |
0.1213998461 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 10 |
Hb_001123_330 |
0.1216173826 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 11 |
Hb_002790_010 |
0.122017634 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 12 |
Hb_000527_020 |
0.1223800322 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 13 |
Hb_001190_030 |
0.1234969443 |
- |
- |
hypothetical protein VITISV_017115 [Vitis vinifera] |
| 14 |
Hb_000654_060 |
0.1258141257 |
- |
- |
hypothetical protein CICLE_v10027550mg [Citrus clementina] |
| 15 |
Hb_125781_010 |
0.1265094945 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_004774_080 |
0.1270490964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_022465_010 |
0.1325518345 |
- |
- |
hypothetical protein JCGZ_06012 [Jatropha curcas] |
| 18 |
Hb_078448_010 |
0.1341782239 |
- |
- |
hypothetical protein L484_017526 [Morus notabilis] |
| 19 |
Hb_003680_180 |
0.1352953778 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 20 |
Hb_004092_020 |
0.136901844 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 32 [Jatropha curcas] |