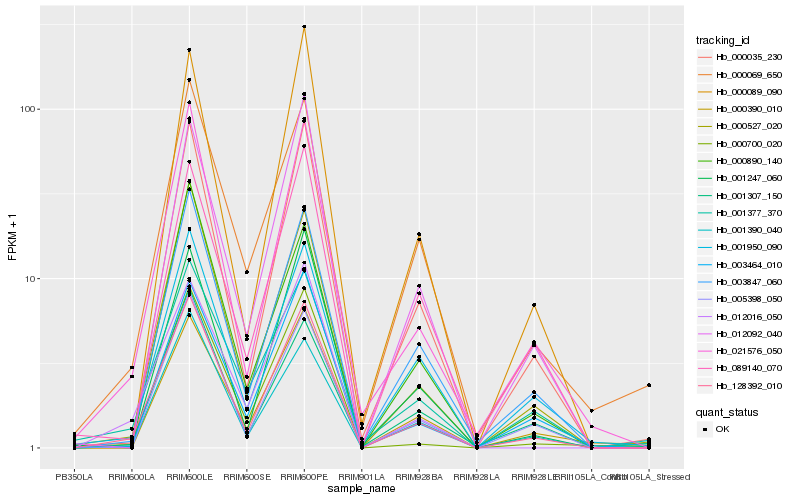
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_128392_010 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 2 |
Hb_021576_050 |
0.074694165 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 3 |
Hb_000527_020 |
0.0816665372 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 4 |
Hb_000035_230 |
0.0828912821 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 5 |
Hb_003847_060 |
0.0844109141 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 6 |
Hb_005398_050 |
0.0990219679 |
- |
- |
PREDICTED: uncharacterized protein LOC105644215 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003464_010 |
0.1026283791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000069_650 |
0.1071676738 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001247_060 |
0.1100432421 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_001950_090 |
0.1102861877 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 11 |
Hb_012092_040 |
0.1157130961 |
- |
- |
PREDICTED: expansin-A1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_012016_050 |
0.118466947 |
- |
- |
hypothetical protein JCGZ_09036 [Jatropha curcas] |
| 13 |
Hb_001377_370 |
0.1193041241 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 14 |
Hb_000089_090 |
0.1193295141 |
- |
- |
hypothetical protein POPTR_0019s12670g [Populus trichocarpa] |
| 15 |
Hb_000700_020 |
0.1194407979 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 16 |
Hb_001307_150 |
0.1222769522 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 17 |
Hb_001390_040 |
0.122964572 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 18 |
Hb_000390_010 |
0.1236234276 |
- |
- |
PREDICTED: protein CDI [Jatropha curcas] |
| 19 |
Hb_089140_070 |
0.1253120077 |
- |
- |
PREDICTED: uncharacterized protein LOC105637961 [Jatropha curcas] |
| 20 |
Hb_000890_140 |
0.1257698442 |
- |
- |
PREDICTED: uncharacterized protein LOC105646185 [Jatropha curcas] |