| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
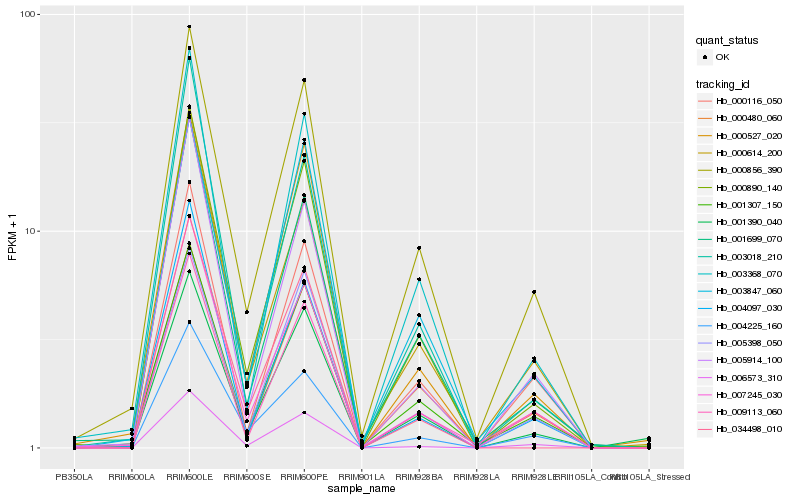
Hb_000890_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646185 [Jatropha curcas] |
| 2 |
Hb_000527_020 |
0.0754686732 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 3 |
Hb_003368_070 |
0.0758628402 |
- |
- |
PREDICTED: uncharacterized protein LOC105109387 [Populus euphratica] |
| 4 |
Hb_001699_070 |
0.0776406444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000856_390 |
0.0823623317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007245_030 |
0.0823856307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_034498_010 |
0.0868820904 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 8 |
Hb_005398_050 |
0.0900786113 |
- |
- |
PREDICTED: uncharacterized protein LOC105644215 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001390_040 |
0.0911628789 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 10 |
Hb_003847_060 |
0.0935350914 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 11 |
Hb_006573_310 |
0.0973189866 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 12 |
Hb_001307_150 |
0.0980496465 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 13 |
Hb_003018_210 |
0.1001490905 |
- |
- |
PREDICTED: uncharacterized protein LOC105138721 isoform X2 [Populus euphratica] |
| 14 |
Hb_004225_160 |
0.1009792001 |
- |
- |
PREDICTED: kiwellin-like [Jatropha curcas] |
| 15 |
Hb_005914_100 |
0.1025234688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000116_050 |
0.1047441093 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000614_200 |
0.1049272009 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 18 |
Hb_004097_030 |
0.1057448931 |
- |
- |
PREDICTED: uncharacterized protein LOC105634518 [Jatropha curcas] |
| 19 |
Hb_009113_060 |
0.1061224173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000480_060 |
0.1107007262 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |