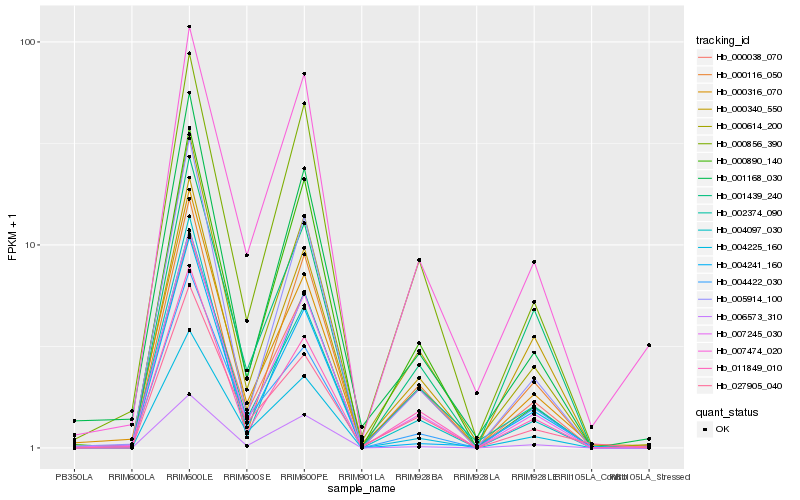
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004225_160 |
0.0 |
- |
- |
PREDICTED: kiwellin-like [Jatropha curcas] |
| 2 |
Hb_007245_030 |
0.0710809949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000038_070 |
0.0881167605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004422_030 |
0.0881517369 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK7-like [Jatropha curcas] |
| 5 |
Hb_000614_200 |
0.0894058043 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 6 |
Hb_006573_310 |
0.0906553023 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 7 |
Hb_027905_040 |
0.0979136249 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: E2F transcription factor-like E2FF [Jatropha curcas] |
| 8 |
Hb_000316_070 |
0.0979625177 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 9 |
Hb_000890_140 |
0.1009792001 |
- |
- |
PREDICTED: uncharacterized protein LOC105646185 [Jatropha curcas] |
| 10 |
Hb_000856_390 |
0.1016597628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_011849_010 |
0.1017179504 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 12 |
Hb_005914_100 |
0.1045707247 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001439_240 |
0.1069897188 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0017s12230g [Populus trichocarpa] |
| 14 |
Hb_002374_090 |
0.1115893584 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 15 |
Hb_007474_020 |
0.1118533025 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 16 |
Hb_004241_160 |
0.1154171222 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-7 [Jatropha curcas] |
| 17 |
Hb_000116_050 |
0.1170401317 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_001168_030 |
0.1178468922 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 19 |
Hb_004097_030 |
0.117949678 |
- |
- |
PREDICTED: uncharacterized protein LOC105634518 [Jatropha curcas] |
| 20 |
Hb_000340_550 |
0.1189361543 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |